Molecular Insights into Host and Vector Manipulation by Plant Viruses
Abstract
:1. Introduction
2. The Interplay between CMV 2b and JA Signalling
3. Potyviruses: An Intriguing Strategy of Protein Relocalisation to Manipulate Aphids
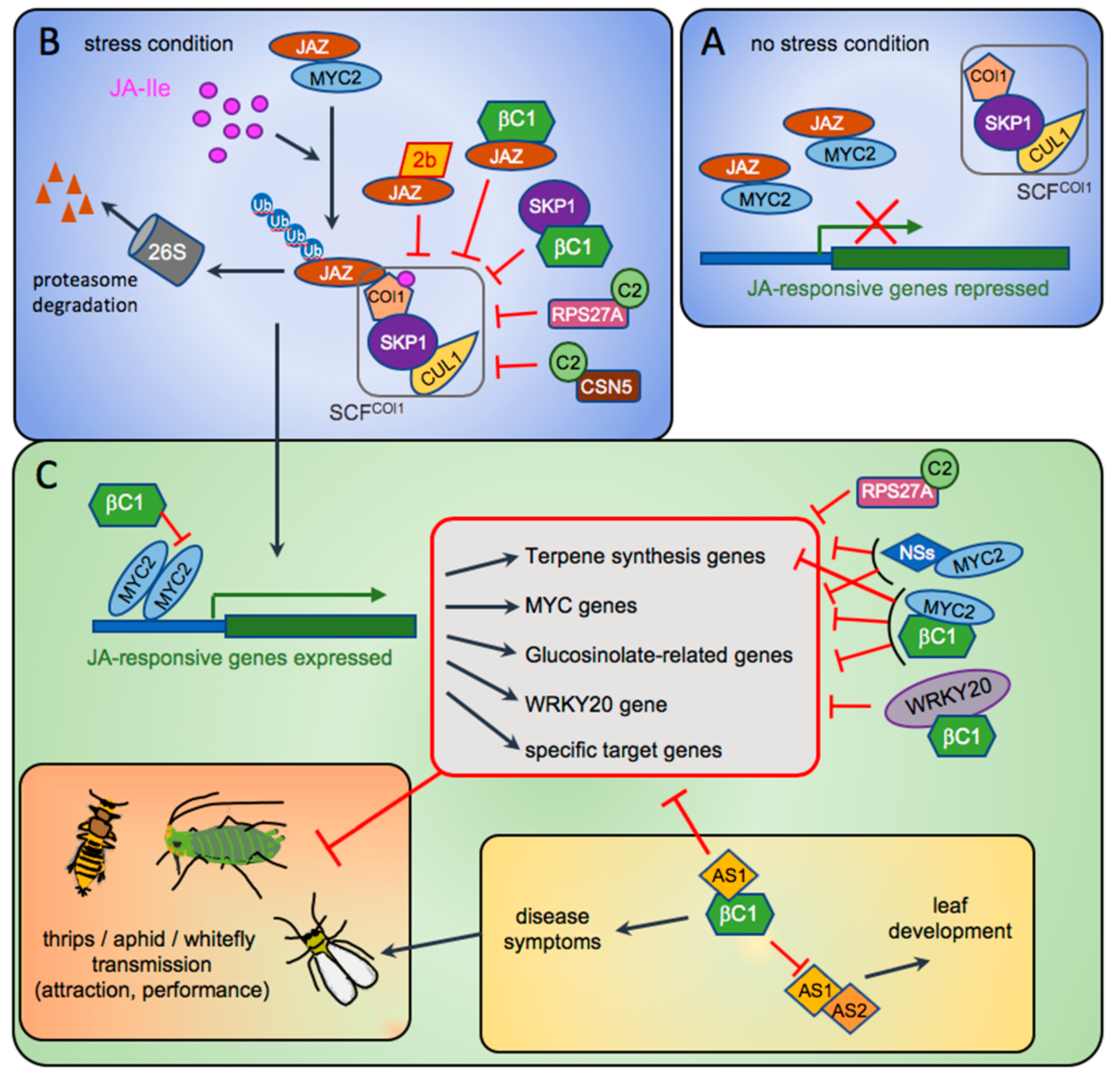
4. Geminiviruses: Master Inventors of Outstanding Pathogenic Factors
5. Tospovirus: One More Virus Playing with JA Signalling
6. Luteoviridae: Still a Puzzling Situation
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, J.C.K.; Falk, B.W. Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annu. Rev. Phytopathol. 2006, 44, 183–212. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brault, V.; Uzest, M.; Monsion, B.; Jacquot, E.; Blanc, S. Aphids as transport devices for plant viruses. C. R. Biol. 2010, 333, 524–538. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant Virus – Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef]
- Gray, S.; Cilia, M.; Ghanim, M. Circulative, “ Nonpropagative ” Virus Transmission: An Orchestra Plant-Derived Instruments. Adv. Virus Res 2014, 89, 141–199. [Google Scholar]
- Deshoux, M.; Monsion, B.; Uzest, M. Insect cuticular proteins and their role in transmission of phytoviruses. Curr. Opin. Virol. 2018, 33, 137–143. [Google Scholar] [CrossRef]
- Blanc, S.; Michalakis, Y. Manipulation of hosts and vectors by plant viruses and impact of the environment. Curr. Opin. Insect Sci. 2016, 16, 36–43. [Google Scholar] [CrossRef]
- Dáder, B.; Then, C.; Berthelot, E.; Ducousso, M.; Ng, J.C.K.; Drucker, M. Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Sci. 2017, 24, 929–946. [Google Scholar] [CrossRef]
- Mauck, K.E. Variation in virus effects on host plant phenotypes and insect vector behavior: What can it teach us about virus evolution? Curr. Opin. Virol. 2016, 21, 114–123. [Google Scholar] [CrossRef]
- Mauck, K.E.; Chesnais, Q.; Shapiro, L.R. Evolutionary Determinants of Host and Vector Manipulation by Plant Viruses. Adv. Virus Res. 2018, 101, 189–250. [Google Scholar] [PubMed]
- Carr, J.P.; Donnelly, R.; Tungadi, T.; Murphy, A.M.; Jiang, S.; Bravo-Cazar, A.; Yoon, J.Y.; Cunniffe, N.J.; Glover, B.J.; Gilligan, C.A. Viral Manipulation of Plant Stress Responses and Host Interactions With Insects. Adv. Virus Res. 2018, 102, 177–197. [Google Scholar] [PubMed]
- Eigenbrode, S.D.; Bosque-Pérez, N.A.; Davis, T.S. Insect-Borne Plant Pathogens and Their Vectors: Ecology, Evolution, and Complex Interactions. Annu. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Fereres, A.; Moreno, A. Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Res. 2009, 141, 158–168. [Google Scholar] [CrossRef]
- Shalileh, S.; Ogada, P.A.; Moualeu, D.P.; Poehling, H. Behavior Manipulation of Frankliniella occidentalis (Thysanoptera: Thripidae) by Tomato Spotted Wilt Virus (Tospovirus) Via the Host Plant Nutrients to Enhance Its Transmission and Spread. Environ. Entomol. 2016, 45, 1235–1242. [Google Scholar] [CrossRef] [Green Version]
- Ingwell, L.L.; Eigenbrode, S.D.; Bosque-Pérez, N.A. Plant viruses alter insect behavior to enhance their spread. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Rajabaskar, D.; Bosque-Pérez, N.A.; Eigenbrode, S.D. Preference by a virus vector for infected plants is reversed after virus acquisition. Virus Res. 2014, 186, 32–37. [Google Scholar] [CrossRef]
- Mauck, K.; Bosque-Pérez, N.A.; Eigenbrode, S.D.; De Moraes, C.M.; Mescher, M.C. Transmission mechanisms shape pathogen effects on host-vector interactions: Evidence from plant viruses. Funct. Ecol. 2012, 26, 1162–1175. [Google Scholar] [CrossRef]
- Jacquemond, M. Cucumber Mosaic Virus, Academic Press: Cambridge, MA, USA, 2012; 84, 439–504.
- Palukaitis, P.; Garcia-Arenal, F. Cucumoviruses. Adv. Virus. Res. 2003, 62, 241–323. [Google Scholar]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef] [Green Version]
- Carmo-Sousa, M.; Moreno, A.; Garzo, E.; Fereres, A. A non-persistently transmitted-virus induces a pull-push strategy in its aphid vector to optimize transmission and spread. Virus Res. 2014, 186, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Pantaleo, V.; Burgyan, J. RNA silencing: An antiviral mechanism. Adv. Virus. Res. 2009, 75, 35–71. [Google Scholar]
- Zhao, J.H.; Liu, X.L.; Fang, Y.Y.; Fang, R.X.; Guo, H.S. CMV2b-Dependent Regulation of Host Defense Pathways in the Context of Viral Infection. Viruses 2018, 10, 618. [Google Scholar] [CrossRef] [Green Version]
- Lewsey, M.G.; Murphy, A.M.; Maclean, D.; Dalchau, N.; Westwood, J.H.; Macaulay, K.; Bennett, M.H.; Moulin, M.; Hanke, D.E.; Powell, G.; et al. Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol. Plant Microbe Interact 2010, 23, 835–845. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Jander, G. Plant Immunity to Insect Herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [Green Version]
- Ziebell, H.; Murphy, A.M.; Groen, S.C.; Tungadi, T.; Westwood, J.H.; Lewsey, M.G.; Moulin, M.; Kleczkowski, A.; Smith, A.G.; Stevens, M.; et al. Cucumber mosaic virus and its 2b RNA silencing suppressor modify plant-aphid interactions in tobacco. Sci. Rep. 2011, 1, 1–7. [Google Scholar] [CrossRef]
- Wu, D.; Qi, T.; Li, W.X.; Tian, H.; Gao, H.; Wang, J.; Ge, J.; Yao, R.; Ren, C.; Wang, X.B.; et al. Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 2017, 27, 402–415. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites — Pathways, transcription factors and applied aspects — A brief review. N. Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tungadi, T.; Groen, S.C.; Murphy, A.M.; Pate, A.E.; Iqbal, J.; Bruce, T.J.A.; Cunniffe, N.J.; Carr, J.P. Cucumber mosaic virus and its 2b protein alter emission of host volatile organic compounds but not aphid vector settling in tobacco. Virol. J. 2017, 14, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tungadi, T.; Donnelly, R.; Qing, L.; Iqbal, J.; Murphy, A.M.; Pate, A.E.; Cunniffe, N.J.; Carr, J.P. Cucumber mosaic virus 2b proteins inhibit virus-induced aphid resistance in tobacco. Mol. Plant Pathol. 2019, 21, 250–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westwood, J.H.; Groen, S.C.; Du, Z.; Murphy, A.M.; Anggoro, D.T.; Tungadi, T.; Luang-In, V.; Lewsey, M.G.; Rossiter, J.T.; Powell, G.; et al. A trio of viral proteins tunes aphid-plant interactions in arabidopsis thaliana. PLoS ONE 2013, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Jander, G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 2007, 49, 1008–1019. [Google Scholar] [CrossRef]
- Revers, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Casteel, C.L.; Yang, C.; Nanduri, A.C.; De Jong, H.N.; Whitham, S.A.; Jander, G. The NIa-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid). Plant J. 2014, 77, 653–663. [Google Scholar] [CrossRef]
- Rajamäki, M.L.; Valkonen, J.P.T. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like potato virus a in nicotiana species. Plant Cell 2009, 21, 2485–2502. [Google Scholar] [CrossRef] [Green Version]
- Anindya, R.; Savithri, H.S. Potyviral NIa proteinase, a proteinase with novel deoxyribonuclease activity. J. Biol. Chem. 2004, 279, 32159–32169. [Google Scholar] [CrossRef] [Green Version]
- Bak, A.; Cheung, A.L.; Yang, C.; Whitham, S.A.; Casteel, C.L. A viral protease relocalizes in the presence of the vector to promote vector performance. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Martinière, A.; Bak, A.; Macia, J.L.; Lautredou, N.; Gargani, D.; Doumayrou, J.; Garzo, E.; Moreno, A.; Fereres, A.; Blanc, S.; et al. A virus responds instantly to the presence of the vector on the host and forms transmission morphs. eLife 2013, 2013, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, E.; Ducousso, M.; Macia, J.-L.; Bogaert, F.; Baecker, V.; Thébaud, G.; Gallet, R.; Yvon, M.; Blanc, S.; Khelifa, M.; et al. Turnip Mosaic Virus Is a Second Example of a Virus Using Transmission Activation for Plant-to-Plant Propagation by Aphids. J. Virol. 2019, 93, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthelot, E.; Macia, J.L.; Martinière, A.; Morisset, A.; Gallet, R.; Blanc, S.; Khelifa, M.; Drucker, M. Pharmacological analysis of transmission activation of two aphid-vectored plant viruses, turnip mosaic virus and cauliflower mosaic virus. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Casteel, C.L.; De Alwis, M.; Bak, A.; Dong, H.; Whitham, S.A.; Jander, G. Disruption of Ethylene Responses by Turnip mosaic virus Mediates Suppression of Plant Defense against the Green Peach Aphid Vector. Plant Physiol. 2015, 169, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Bak, A.; Patton, M.K.F.; Perilla-Henao, L.M.; Aegerter, B.J.; Casteel, C.L. Ethylene signaling mediates potyvirus spread by aphid vectors. Oecologia 2019, 190, 139–148. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging Virus Diseases Transmitted by Whiteflies. Ann. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef]
- Yang, X.; Guo, W.; Li, F.; Sunter, G.; Zhou, X. Geminivirus-Associated Betasatellites: Exploiting Chinks in the Antiviral Arsenal of Plants. Trends Plant Sci. 2019, 24, 519–529. [Google Scholar] [CrossRef]
- Morin, S.; Ghanim, M.; Zeidan, M.; Czosnek, H.; Verbeek, M.; Van Den Heuvel, J.F.J.M. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology 1999, 256, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.V. Plant antiviral immunity against geminiviruses and viral counter-defense for survival. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyan, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnanasekaran, P.; KishoreKumar, R.; Bhattacharyya, D.; Vinoth Kumar, R.; Chakraborty, S. Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol. Plant Pathol. 2019, 20, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, X.; Bisaro, D.M.; Zhou, X. The βC1 Protein of Geminivirus–Betasatellite Complexes: A Target and Repressor of Host Defenses. Mol. Plant 2018, 11, 1424–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.H. βCl, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [Green Version]
- Nurmberg, P.L.; Knox, K.A.; Yun, B.W.; Morris, P.C.; Shafiei, R.; Hudson, A.; Loake, G.J. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 18795–18800. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Luan, J.B.; Qi, J.F.; Huang, C.J.; Li, M.; Zhou, X.P.; Liu, S.S. Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 2012, 21, 1294–1304. [Google Scholar] [CrossRef]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, I.T.; Kessler, A.; Halitschke, R. Volatile signaling in plant–plant–herbivore interactions: What is real? Curr Opin Plant Biol 2002, 5, 351–354. [Google Scholar] [CrossRef]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J.A. Role of Glucosinolates in Insect-Plant Relationships and Multitrophic Interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef]
- Rosas-Díaz, T.; Macho, A.P.; Beuzón, C.R.; Lozano-Durán, R.; Bejarano, E.R. The C2 protein from the geminivirus Tomato Yellow leaf curl sardinia virus decreases sensitivity to jasmonates and suppresses jasmonate-mediated defences. Plants 2016, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, C.; Deng, W.H.; Yao, D.M.; Pan, L.L.; Li, Y.Q.; Liu, Y.Q.; Liang, Y.; Zhou, X.P.; Wang, X.W. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, J.B.; Wang, X.W.; Colvin, J.; Liu, S.S. Plant-mediated whitefly-begomovirus interactions: Research progress and future prospects. Bull. Entomol. Res. 2014, 104, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.; Selth, L.A.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA β satellite. Mol. Plant-Microbe Interact. 2009, 22, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Hu, T.; Bao, M.; Cao, L.; Zhang, H.; Song, F.; Xie, Q.; Zhou, X. Tobacco RING E3 Ligase NtRFP1 Mediates Ubiquitination and Proteasomal Degradation of a Geminivirus-Encoded βC1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, 1–30. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co- option of SCF-mediated ubiquitination. Plant Signal Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Yao, X.; Cai, C.; Li, R.; Du, J.; Sun, Y.; Wang, M.; Zou, Z.; Wang, Q.; Kliebenstein, D.J.; et al. Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci. Adv. 2019, 5, eaav9801. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, A.E.; Ullman, D.E.; German, T.L. Tospovirus-Thrips Interactions. Annu. Rev. Phytopathol. 2005, 43, 459–489. [Google Scholar] [CrossRef]
- Stafford, C.A.; Walker, G.P.; Ullman, D.E. Infection with a plant virus modifies vector feeding behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 9350–9355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogada, P.A.; Maiss, E.; Poehling, H.M. Influence of tomato spotted wilt virus on performance and behaviour of western flower thrips (Frankliniella occidentalis). J. Appl. Entomol. 2013, 137, 488–498. [Google Scholar] [CrossRef]
- Abe, H.; Tomitaka, Y.; Shimoda, T.; Seo, S.; Sakurai, T.; Kugimiya, S.; Tsuda, S.; Kobayashi, M. Antagonistic plant defense system regulated by phytohormones assists interactions among vector insect, thrips and a tospovirus. Plant Cell Physiol. 2012, 53, 204–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Xu, S.; Zhao, P.; Zhang, X.; Yao, X.; Sun, Y.; Fang, R.; Ye, J. The orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog. 2019, 15, e1007897. [Google Scholar] [CrossRef] [PubMed]
- Hedil, M.; Sterken, M.G.; De Ronde, D.; Lohuis, D.; Kormelink, R. Analysis of tospovirus nss proteins in suppression of systemic silencing. PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Gildow, F.E. Luteovirus–aphid interactions. Annu. Rev. Phytopathol. 2003, 41, 539–566. [Google Scholar] [CrossRef] [PubMed]
- Bosque-Pérez, N.A.; Eigenbrode, S.D. The influence of virus-induced changes in plants on aphid vectors: Insights from luteovirus pathosystems. Virus Res. 2011, 159, 201–205. [Google Scholar] [CrossRef]
- Eigenbrode, S.D.; Ding, H.; Shiel, P.; Berger, P.H. Volatiles from potato plants infected with potato leafroll virus attract and arrest the virus vector, Myzus persicae Homoptera: Aphididae. Proc. Biol. Sci. 2002, 269, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Medina-Ortega, K.J.; Bosque-Pérez, N.A.; Ngumbi, E.; Jiménez-Martínez, E.S.; Eigenbrode, S.D. Rhopalosiphum padi (Hemiptera: Aphididae) responses to volatile cues from Barley yellow dwarf virus-infected wheat. Environ. Entomol. 2009, 38, 836–845. [Google Scholar] [CrossRef] [Green Version]
- Claudel, P.; Chesnais, Q.; Fouché, Q.; Krieger, C.; Halter, D.; Bogaert, F.; Meyer, S.; Boissinot, S.; Hugueney, P.; Ziegler-Graff, V.; et al. The Aphid-Transmitted Turnip yellows virus Differentially Affects Volatiles Emission and Subsequent Vector Behavior in Two Brassicaceae Plants. Int. J. Mol. Sci. 2018, 19, 2316. [Google Scholar] [CrossRef] [Green Version]
- Werner, B.J.; Mowry, T.M.; Bosque-Perez, N.A.; Ding, H.; Eigenbrode, S.D. Changes in green peach aphid responses to potato leafroll virus-induced volatiles emitted during disease progression. Env. Entomol. 2009, 38, 1429–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmo-Sousa, M.; Moreno, A.; Plaza, M.; Garzo, E.; Fereres, A. Cucurbit aphid-borne yellows virus (CABYV) modifies the alighting, settling and probing behaviour of its vector Aphis gossypii favouring its own spread. Ann. Appl. Biol. 2016, 169, 284–297. [Google Scholar] [CrossRef]
- Patton, M.F.; Bak, A.; Sayre, J.M.; Heck, M.L.; Casteel, C.L. A polerovirus, Potatao leafrollvirus, alters plant-vector ineractions using three viral proteins. Plant Cell Env. 2019, 1–13. [Google Scholar]
- Pazhouhandeh, M.; Dieterle, M.; Marrocco, K.; Lechner, E.; Berry, B.; Brault, V.; Hemmer, O.; Kretsch, T.; Richards, K.E.; Genschik, P.; et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 2006, 103, 1994–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortolamiol, D.; Pazhouhandeh, M.; Marrocco, K.; Genschik, P.; Ziegler-Graff, V. The Polerovirus F Box Protein P0 Targets ARGONAUTE1 to Suppress RNA Silencing. Curr. Biol. 2007, 17, 1615–1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946. [Google Scholar] [CrossRef] [Green Version]
- Ashoub, A.; Rohde, W.; Prufer, D. In planta transcription of a second subgenomic RNA increases the complexity of the subgroup 2 luteovirus genome. Nucleic Acids Res 1998, 26, 420–426. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef]
- Guo, H.; Gu, L.; Liu, F.; Chen, F.; Ge, F.; Sun, Y. Aphid-borne viral spread is enhanced by virus-induced accumulation of plant reactive oxygen species. Plant Physiol. 2019, 179, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Mauck, K.E.; Kenney, J.; Chesnais, Q. Progress and challenges in identifying molecular mechanisms underlying host and vector manipulation by plant viruses. Curr. Opin. Insect Sci. 2019, 7–18. [Google Scholar] [CrossRef]
- Westwood, J.H.; Lewsey, M.G.; Murphy, A.M.; Tungadi, T.; Bates, A.; Gilligan, C.A.; Carr, J.P. Interference with jasmonic acid-regulated gene expression is a general property of viral suppressors of RNA silencing but only partly explains virus-induced changes in plant-aphid interactions. J. Gen. Virol. 2014, 95, 733–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Kügler, A.; McGale, E.; Haverkamp, A.; Knaden, M.; Guo, H.; Beran, F.; Yon, F.; Li, R.; Lackus, N.; et al. Tissue-Specific Emission of (E)-α-Bergamotene Helps Resolve the Dilemma When Pollinators Are Also Herbivores. Curr. Biol. 2017, 27, 1336–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Ye, J. Manipulation of jasmonate signaling by plant viruses and their insect vectors. Viruses 2020, 12, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziegler-Graff, V. Molecular Insights into Host and Vector Manipulation by Plant Viruses. Viruses 2020, 12, 263. https://doi.org/10.3390/v12030263
Ziegler-Graff V. Molecular Insights into Host and Vector Manipulation by Plant Viruses. Viruses. 2020; 12(3):263. https://doi.org/10.3390/v12030263
Chicago/Turabian StyleZiegler-Graff, Véronique. 2020. "Molecular Insights into Host and Vector Manipulation by Plant Viruses" Viruses 12, no. 3: 263. https://doi.org/10.3390/v12030263
APA StyleZiegler-Graff, V. (2020). Molecular Insights into Host and Vector Manipulation by Plant Viruses. Viruses, 12(3), 263. https://doi.org/10.3390/v12030263




