SOX2 Represses Hepatitis B Virus Replication by Binding to the Viral EnhII/Cp and Inhibiting the Promoter Activation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Study
2.2. Plasmids and Constructions
2.3. Cell Culture and Transfection
2.4. Quantitative RT-PCR Analysis
2.5. Western Blot
2.6. HBV DNA Quantification
2.7. Northern Blot
2.8. Dual-Luciferase Reporter Assay
2.9. Nuclear and Cytoplasmic Extraction Reagents
2.10. Chromatin Immunoprecipitation (ChIP) Assay
2.11. Immunofluorescence and Confocal Analysis
2.12. Statistical Analysis
3. Results
3.1. HBV Induces SOX2 Expression
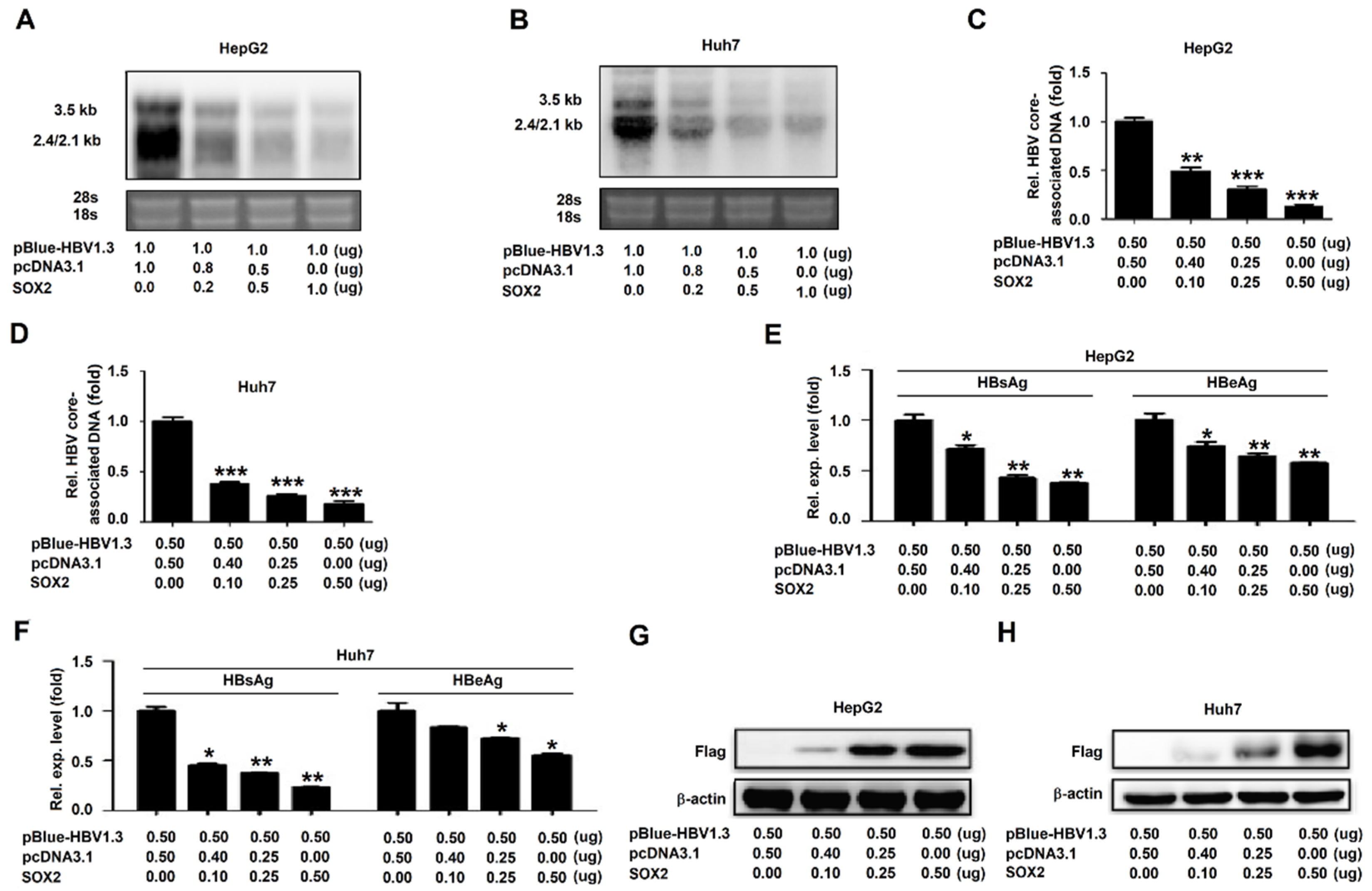
3.2. SOX2 Represses HBV Replication in HepG2 Cells and Huh7 Cells
3.3. SOX2 Represses HBV Replication through Inhibiting EnhII/Cp Activation
3.4. The HMG Domain is Required for SOX2-Mediated Repression of HBV Replication
3.5. SOX2 Suppresses HBV Replication in BALB/c Mice
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol. 2018, 3, 383–403. [Google Scholar] [CrossRef]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.W.; Kirchner, J.T. Hepatitis B. Am. Fam. Physician. 2004, 69, 75–82. [Google Scholar] [PubMed]
- Chang, M.H. Hepatitis B virus infection. Semin. Fetal Neonatal. Med. 2007, 12, 160–167. [Google Scholar] [CrossRef]
- Schlicht, H.J.; Galle, P.; Schaller, H. The hepatitis-B viruses: Molecular biology and recent tissue culture systems. J. Cell Sci. Suppl. 1987, 7, 197–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kann, M.; Schmitz, A.; Rabe, B. Intracellular transport of hepatitis B virus. World J. Gastroenterol. 2007, 13, 39–47. [Google Scholar] [CrossRef]
- Ezzikouri, S.; Ozawa, M.; Kohara, M.; Elmdaghri, N.; Benjelloun, S.; Tsukiyama-Kohara, K. Recent insights into hepatitis B virus-host interactions. J. Med. Virol. 2014, 86, 925–932. [Google Scholar] [CrossRef]
- Kramvis, A.; Kew, M.C. The core promoter of hepatitis B virus. J. Viral. Hepat. 1999, 6, 415–427. [Google Scholar] [CrossRef]
- Nishitsuji, H.; Ujino, S.; Harada, K.; Shimotohno, K. TIP60 complex inhibits HBV transcription. J. Virol. 2018, 92. pii: e01788-17. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Qiu, J.; Li, B.; Hong, J.; Lu, C.; Wang, L.; Wang, J.; Hu, Y.; Jia, W.; Yuan, Y. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin. Biochem. 2011, 44, 582–589. [Google Scholar] [CrossRef]
- Carrasco-García, E.; Moreno-Cugnon, L.; Garcia, I.; Borras, C.; Revuelta, M.; Izeta, A.; Lopez-Lluch, G.; Pancorbo, M.; Vergara, I.; Vina, J.; et al. SOX2 expression diminishes with ageing in several tissues in mice and humans. Mech. Ageing Dev. 2018, 177. [Google Scholar]
- Liu, L.; Liu, C.; Zhang, Q.; Shen, J.; Zhang, H.; Shan, J.; Duan, G.; Guo, D.; Chen, X.; Cheng, J.; et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells: Regulation of liver cancer stem cells by SIRT1. Hepatology 2016, 64, 814–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, F.; Cheng, X.; Sun, S.; Zhou, J. Transcriptional activation of PD-L1 by Sox2 contributes to the proliferation of hepatocellular carcinoma cells. Oncol. Rep. 2017, 37, 3061–3067. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, Y.; Zhang, M.; Yang, Y.; Chang, L. miR-126 inhibits cell proliferation and induces cell apoptosis of hepatocellular carcinoma cells partially by targeting Sox2. Hum. Cell. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dai, X.; Wang, T.; Zhang, C.; Zhang, W.; Zhang, W.; Zhang, Q.; Wu, K.; Liu, F.; Liu, Y.; et al. Hepatitis B virus PreS1 facilitates hepatocellular carcinoma development by promoting appearance and self-renewal of liver cancer stem cells. Cancer Lett. 2017, 400, 149–160. [Google Scholar] [CrossRef]
- Zhu, M.; Li, W.; Lu, Y.; Dong, X.; Lin, B.; Chen, Y.; Xueer, Z.; Guo, J.; Li, M. HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway: Alpha-fetoprotein promotes HCC stem cell reprogramming. Int. J. Cancer 2016, 140. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, S.; Zhou, Y.; Yang, L.; Zhou, D.; Yang, Y.; Lu, M.; Yang, D.; Song, J. The dose of HBV genome contained plasmid has a great impact on HBV persistence in hydrodynamic injection mouse model. Virol. J. 2017, 14, 205. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Xu, W.; Chen, Y.; Xie, X.; Zhang, Y.; Ma, C.; Yang, Q.; Han, Y.; Zhu, C.; Xiong, Y.; et al. Matrix metalloproteinase 9 facilitates hepatitis B virus replication through binding with type I interferon (IFN) receptor 1 to repress IFN/JAK/STAT signaling. J. Virol. 2017, 91. pii: e01824-16. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Ma, C.; Zhang, Q.; Zhao, R.; Hu, D.; Zhang, X.; Chen, J.; Liu, F.; Wu, K.; Liu, Y.; et al. PJA1 coordinates with the SMC5/6 complex to restrict DNA viruses and episomal genes through interferon-independent manner. J. Virol. 2018, 92. pii: e00825-18. [Google Scholar] [CrossRef] [Green Version]
- Aboushousha, T.; Mamdouh, S.; Hamdy, H.; Helal, N.; Khorshed, F.; Safwat, G.; Sel eem, M. Immunohistochemical and biochemical expression patterns of TTF-1, RAGE, GLUT-1 and SOX2 in HCV-associated hepatocellular carcinomas. Asian Pac. J. Cancer Prev 2018, 19, 219–227. [Google Scholar]
- Wong, D.K.; Yuen, M.F.; Poon, R.T.; Yuen, J.C.; Fung, J.; Lai, C.L. Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. J. Hepatol. 2006, 45, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Halgand, B.; Desterke, C.; Riviere, L.; Fallot, G.; Sebagh, M.; Calderaro, J.; Bioulac-Sage, P.; Neuveut, C.; Buendia, M.A.; Samuel, D.; et al. Hepatitis B Virus Pregenomic RNA in Hepatocellular Carcinoma: A Nosological and Prognostic Determinant. Hepatology (Baltimore, Md.) 2018, 67, 86–96. [Google Scholar] [CrossRef]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral. Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kang, H.; Kim, K.-H. Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J. Gastroenterol. 2016, 22, 7017. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Li, W.; Ren, J.; Huang, Y.; Huang, Y.; Hu, Q.; Chen, J.; Chen, W. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget 2016, 7. [Google Scholar] [CrossRef]
- Wang, W.X.; Li, M.; Wu, X.; Wang, Y.; Li, Z.P. HNF1 is critical for the liver-specific function of HBV enhancer II. Res. Virol. 1998, 149, 99–108. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, J.; Ou, J.H. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J. Virol. 2004, 78, 6908–6914. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xie, Y.; Wu, X.; Kong, Y.; Wang, Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology 1995, 214, 371–378. [Google Scholar] [CrossRef]
- Gilbert, S.; Galarneau, L.; Lamontagne, A.; Roy, S.; Bélanger, L. The hepatitis B virus core promoter is strongly activated by the liver nuclear receptor fetoprotein transcription factor or by ectopically expressed steroidogenic factor 1. J. Virol. 2000, 74, 5032–5039. [Google Scholar] [CrossRef]
- Shang, J.; Zheng, Y.; Guo, X.; Mo, J.; Xie, X.; Xiong, Y.; Liu, Y.; Wu, K.; Wu, J. Hepatitis B virus replication and sex-determining region Y box 4 production are tightly controlled by a novel positive feedback mechanism. Sci. Rep. 2015, 5, 10066. [Google Scholar] [CrossRef] [Green Version]
- Mansouri, S.; Nejad, R.; Karabork, M.; Ekinci, C.; Solaroglu, I.; Aldape, K.D.; Zadeh, G. Sox2: regulation of expression and contribution to brain tumors. CNS Oncol. 2016, 5, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Weina, K.; Utikal, J. SOX2 and cancer: current research and its implications in the clinic. Clin. Transl. Med. 2014, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Mo, J.; Xiang, Q.; Zhao, P.; Song, Y.; Yang, G.; Wu, K.; Liu, Y.; Liu, W.; Wu, J. SOX2 Represses Hepatitis B Virus Replication by Binding to the Viral EnhII/Cp and Inhibiting the Promoter Activation. Viruses 2020, 12, 273. https://doi.org/10.3390/v12030273
Yang H, Mo J, Xiang Q, Zhao P, Song Y, Yang G, Wu K, Liu Y, Liu W, Wu J. SOX2 Represses Hepatitis B Virus Replication by Binding to the Viral EnhII/Cp and Inhibiting the Promoter Activation. Viruses. 2020; 12(3):273. https://doi.org/10.3390/v12030273
Chicago/Turabian StyleYang, Hua, Jiayin Mo, Qi Xiang, Peiyi Zhao, Yunting Song, Ge Yang, Kailang Wu, Yingle Liu, Weiyong Liu, and Jianguo Wu. 2020. "SOX2 Represses Hepatitis B Virus Replication by Binding to the Viral EnhII/Cp and Inhibiting the Promoter Activation" Viruses 12, no. 3: 273. https://doi.org/10.3390/v12030273





