Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function
Abstract
1. Introduction
2. Antibodies Induce ADCC Activities through Diverse Innate Effector Cells
3. In Vitro Assays to Assess Antibody-Induced ADCC Activities
3.1. 51Cr Release Assay
3.2. Target Cell Killing Assay
3.3. NK Cell Activation Assay
3.4. ADCC Reporter Bioassay
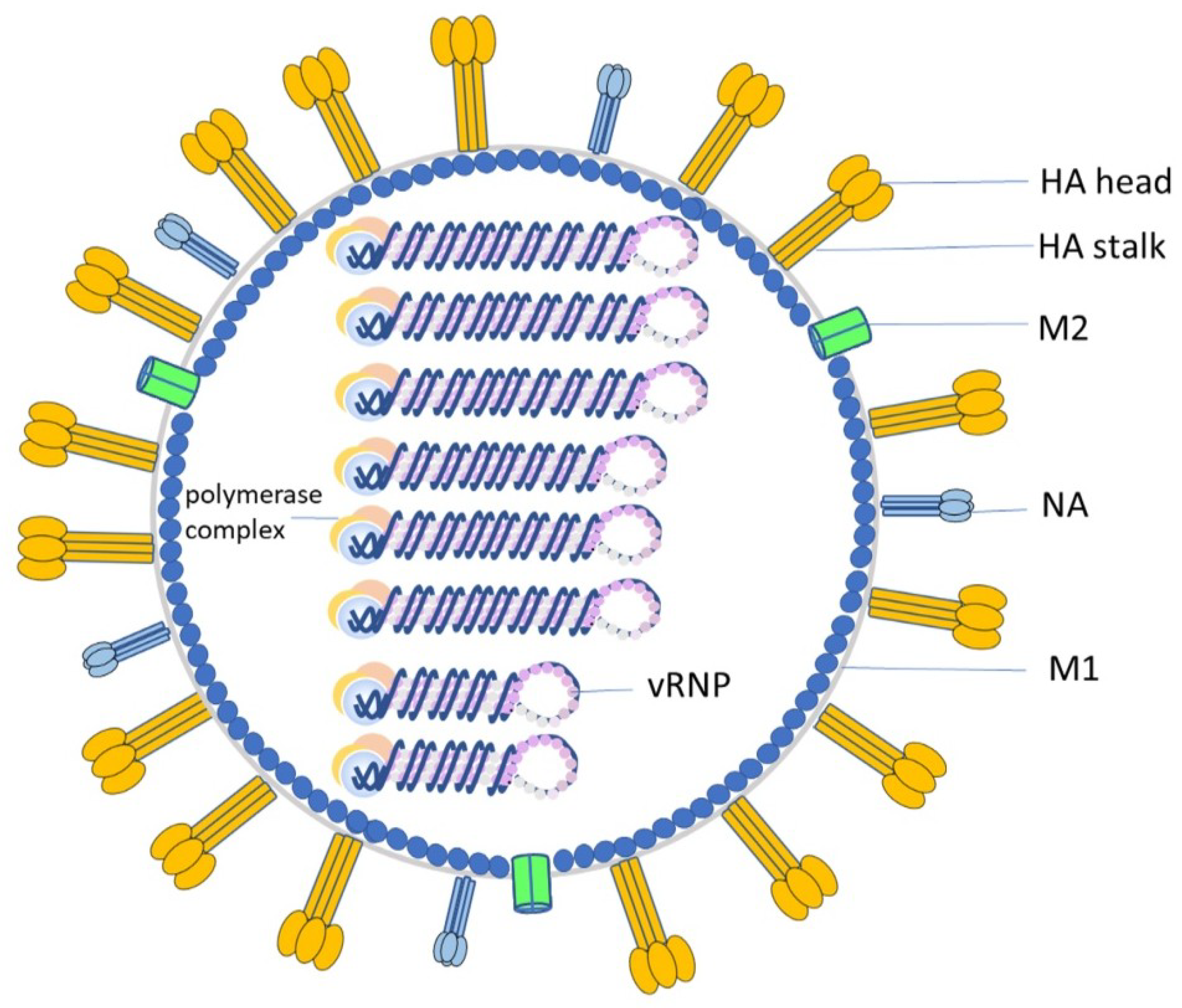
4. ADCC Induced by anti-HA Antibodies
4.1. HA Stalk-Targeting mAbs Can Mediate Robust ADCC Activities
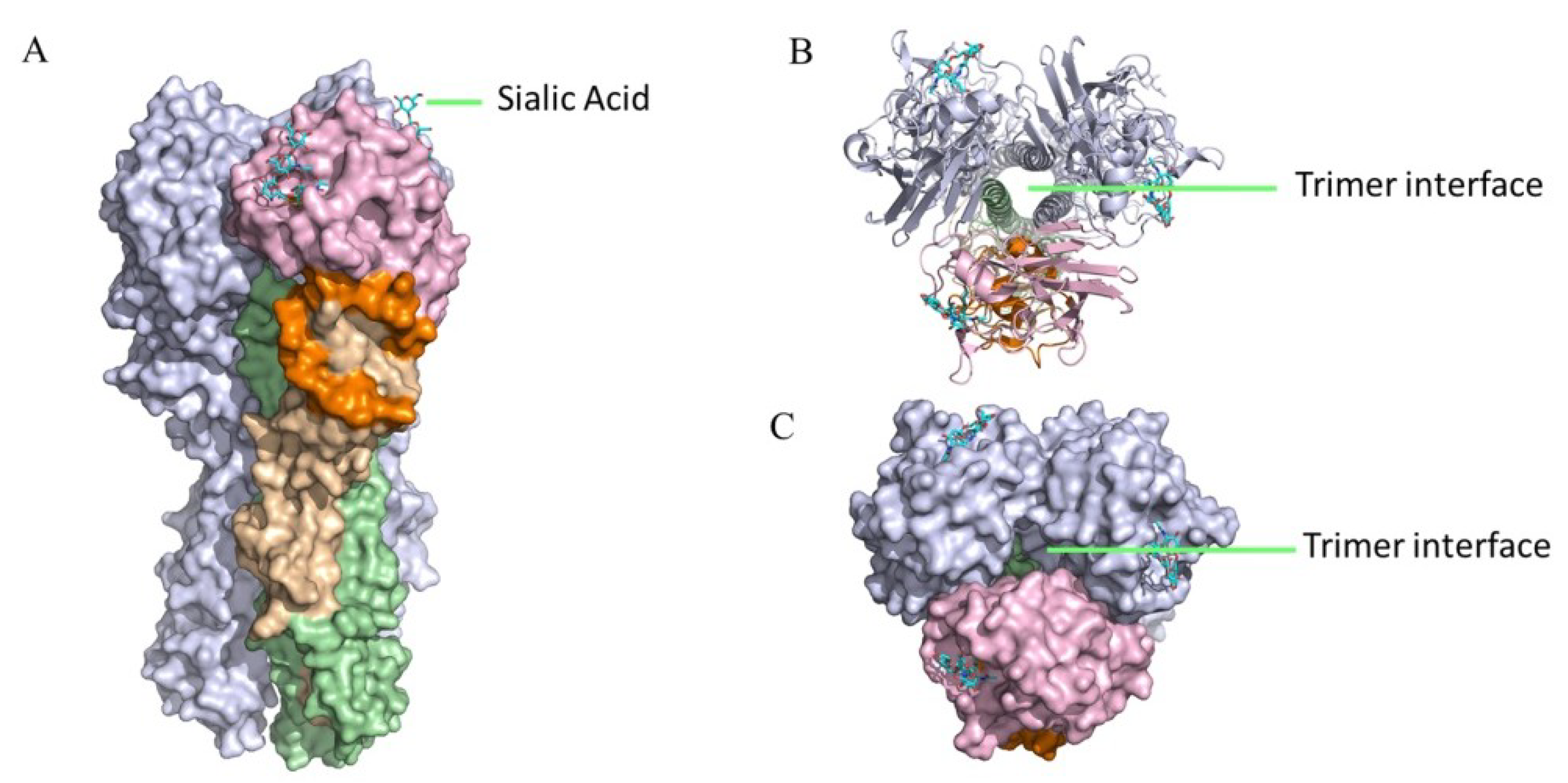
4.2. HI-Negative Head-Targeting mAbs Can Mediate Robust ADCC Activities
4.3. HI-Positive Head-Targeting mAbs Cannot Mediate Robust ADCC Activities

5. ADCC to Other Proteins in Influenza Virus
6. ADCC-Based Vaccine
7. Strategies to Augment ADCC Activity
8. Conclusions and Future Directions
- (1)
- As mAbs mediated Fc-effector activities are epitope dependent, identifying epitopes that can elicit nnAbs will serve as a good start for the rational design of universal vaccines and therapeutics. Some studies show that ADCC-Abs recognize linear epitopes [70]. Arunkumar et al. demonstrated that antibodies recognizing conformational epitopes exhibit better protection compared to the antibodies targeting linear epitopes [17]. Further investigation should be focused on the elucidation of which epitope type can induce a more potent ADCC response and as a result provide better protection.
- (2)
- ADCC-Abs may also exert immune pressure on the virus, resulting in escape mutants that evade antibody recognition similar to what is seen with traditional neutralizing antibodies. No influenza studies have reported ADCC-antibody escape mutants currently, but Chung et al. demonstrated the critical amino acids in the linear epitope could mutate to escape ADCC responses in HIV-1 [139]. Lee et al. summarized two potential mechanisms employed by HIV-1 to resist ADCC responses, i. by directly reducing envelope antigen expression on the infected cell surface and ii. by restricting the exposure of epitopes on the envelope to reduce antibody binding [56,140,141]. Similar or different scenarios may present in influenza, which may need further exploration in future studies.
- (3)
- It remains unclear about the viral replication step(s) in which ADCC occurs. It is indicated that ADCC activity does not intervene in viral entry and fusion and there is a high possibility that ADCC may target the virus-infected cell, which has HA, or other virus protein expressed [142]. This should be better investigated.
- (4)
- Designing antibodies with enhanced capacity to induce ADCC activities against influenza infection should be pursued vigorously. As stated above, new approaches are being developed to augment ADCC in cancer research. These studies in cancer research may provide valuable insights into influenza research towards developing better vaccines or therapeutics to improve influenza patient outcomes. Therefore, it would be beneficial to measure the ADCC potency threshold for protective immunity. However, it is worth noting that a more potent ADCC response may not guarantee better protection. Arunkumar et at. reported that two non-neutralizing antibodies, KL-BHA-9B9 and KL-BHA-4C10, exhibited uncorrelated ADCC activity with in vivo protection [17]. KL-BHA-9B9 had potent ADCC activity in vitro but poor protection in vivo, whereas KL-BHA-4C10 conferred strong protection in vivo but had almost undetectable ADCC in vitro. A better understanding of ADCC activation may facilitate the mechanistic elucidation of this phenomenon as well as other variabilities emerged in current assays.
- (5)
- Although diverse assays have been developed to measure ADCC activity in vitro, it is still obscure to what extent these assays can correlate to the efficacy in vivo. It is challenging to predict ADCC-Ab efficacy in humans. The transgenic mouse model expresses human FcγRs may better evaluate the antibody therapeutic effect in humans. However, mice are not natural hosts of influenza and influenza viruses causing human epidemics or pandemics typically replicate inefficiently in this animal model unless adapted viruses are used. Non-human primate models such as rhesus macaques or cynomolgus macaques are used in ADCC-Abs research and research findings from this model collectively indicated that ADCC-Abs are associated with reduced shedding of influenza viruses [16,130]. Considering the different expression patterns of FcγRs in humans and non-human primate animals, it remains uncertain whether the correlation of treatment outcome can be expected in humans. In this regard, clinical studies regarding ADCC-Abs are instrumental to examine the potential role of ADCC-Abs in combating influenza virus infection in humans [143].
- (6)
- It would be interesting and worthwhile to study the synergy between neutralizing antibody and non-neutralizing antibody in vivo since both classes of antibodies will be elicited after viral infection. The high complexity of each type of antibodies and the lack of an ideal animal model make this investigation relatively challenging. In some cases, the Fc-effector functions induced by nnAb can increase the potency of neutralizing antibodies [14]. In contrast, it is also revealed that Fc-effector functions may not play any role when there is a high dose of neutralizing antibodies [29]. One possible explanation for this is that the function elicited by neutralizing antibodies, such as viral entry blocking or fusion disruption, is potent and sufficient for in vivo protection alone.
- (7)
- The interplay among ADCC-Abs targeting HA, NA, M2e and NP needs to be investigated. It is critical to identify what antibody combination patterns can induce synergistic protection to augment vaccine efficacy. It is reported that the NA-inhibiting antibody elicited weak ADCC response alone, but it may cooperate with antibodies targeting the HA stem to enhance their potency of ADCC activity [83]. NA inhibitors can enhance Fc-dependent functions induced by anti-stalk antibody, suggesting a therapeutic synergy between NA inhibitors and anti-stalk antibody in humans [144]. Deng et al. demonstrated that the vaccination with double-layered nanoparticles containing HA-stalk and M2e induced potent long-lasting immunity and full protection against divergent influenza A viruses challenge in mice, indicating that the combination of antibodies targeting HA stalk domain and M2e can improve the potency and breadth of vaccine-induced protection [145]. The cocktail of therapeutic antibodies may overcome the emergence of viral escape mutants [37]. Further studies on antibody or inhibitor interplay may guide the better design of future universal anti-influenza countermeasures with broad-spectrum protection via multiple mechanisms.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Up to 650,000 People Die of Respiratory Diseases Linked to Seasonal Flu Each Year; WHO: Ginevra, Switzerland, 2017. [Google Scholar]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4), D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Shtyrya, Y.A.; Mochalova, L.V.; Bovin, N.V. Influenza virus neuraminidase: Structure and function. Acta Nat. 2009, 1, 26–32. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef]
- CDC; Weekly, U.S. Influenza Surveillance Report; Key Updates for Week 7, Ending 15 February; CDC: Atlanta, GA, USA, 2020.
- Houser, K.; Subbarao, K. Influenza vaccines: Challenges and solutions. Cell Host Microbe 2015, 17, 295–300. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Alyanak, E.; Broder, K.R.; Walter, E.B.; Fry, A.M.; Jernigan, D.B. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices-United States, 2019–2020 Influenza Season. MMWR Recomm. Rep. 2019, 68, 1–21. [Google Scholar] [CrossRef]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; Garcia-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef]
- Giles, B.M.; Ross, T.M. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 2011, 29, 3043–3054. [Google Scholar] [CrossRef]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortes-Garcia, G.; Vogel, T.U.; et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Reading, P.C.; Kent, S.J. Influenza-specific antibody-dependent cellular cytotoxicity: Toward a universal influenza vaccine. J. Immunol. 2014, 193, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Von Holle, T.A.; Moody, M.A. Influenza and Antibody-Dependent Cellular Cytotoxicity. Front. Immunol. 2019, 10, 1457. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Weinfurter, J.T.; Friedrich, T.C.; Kent, S.J. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J. Virol. 2013, 87, 5512–5522. [Google Scholar] [CrossRef]
- Asthagiri Arunkumar, G.; Ioannou, A.; Wohlbold, T.J.; Meade, P.; Aslam, S.; Amanat, F.; Ayllon, J.; Garcia-Sastre, A.; Krammer, F. Broadly Cross-Reactive, Nonneutralizing Antibodies against Influenza B Virus Hemagglutinin Demonstrate Effector Function-Dependent Protection against Lethal Viral Challenge in Mice. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Henry Dunand, C.J.; Leon, P.E.; Huang, M.; Choi, A.; Chromikova, V.; Ho, I.Y.; Tan, G.S.; Cruz, J.; Hirsh, A.; Zheng, N.Y.; et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016, 19, 800–813. [Google Scholar] [CrossRef]
- Padilla-Quirarte, H.O.; Lopez-Guerrero, D.V.; Gutierrez-Xicotencatl, L.; Esquivel-Guadarrama, F. Protective Antibodies Against Influenza Proteins. Front. Immunol. 2019, 10, 1677. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Luke, C.; Hickman, H.D.; Sangster, M.Y.; Wieland-Alter, W.F.; McBride, J.M.; Yewdell, J.W.; Wright, P.F.; Treanor, J.; Rosenberger, C.M.; et al. Generation and Protective Ability of Influenza Virus-Specific Antibody-Dependent Cellular Cytotoxicity in Humans Elicited by Vaccination, Natural Infection, and Experimental Challenge. J. Infect. Dis. 2016, 214, 945–952. [Google Scholar] [CrossRef]
- de Vries, R.D.; Nieuwkoop, N.J.; Pronk, M.; de Bruin, E.; Leroux-Roels, G.; Huijskens, E.G.W.; van Binnendijk, R.S.; Krammer, F.; Koopmans, M.P.G.; Rimmelzwaan, G.F. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine 2017, 35, 238–247. [Google Scholar] [CrossRef]
- Huber, V.C.; Lynch, J.M.; Bucher, D.J.; Le, J.; Metzger, D.W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001, 166, 7381–7388. [Google Scholar] [CrossRef]
- Zhong, W.; Liu, F.; Wilson, J.R.; Holiday, C.; Li, Z.N.; Bai, Y.; Tzeng, W.P.; Stevens, J.; York, I.A.; Levine, M.Z. Antibody-Dependent Cell-Mediated Cytotoxicity to Hemagglutinin of Influenza A Viruses After Influenza Vaccination in Humans. Open Forum Infect. Dis. 2016, 3, 102. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.; Shin, H.S.; Rhee, S.G. Tyrosine phosphorylation of phospholipase C-gamma 1 induced by cross-linking of the high-affinity or low-affinity Fc receptor for IgG in U937 cells. Proc. Natl. Acad. Sci. USA 1992, 89, 3659–3663. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015, 6, 368. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]
- Salk, H.M.; Haralambieva, I.H.; Ovsyannikova, I.G.; Goergen, K.M.; Poland, G.A. Granzyme B ELISPOT assay to measure influenza-specific cellular immunity. J. Immunol. Methods 2013, 398, 44–50. [Google Scholar] [CrossRef]
- Ravetch, J.V.; Bolland, S. IgG Fc receptors. Annu. Rev. Immunol. 2001, 19, 275–290. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef]
- Weiner, L.M.; Surana, R.; Wang, S. Monoclonal antibodies: Versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010, 10, 317–327. [Google Scholar] [CrossRef]
- Schroeder, H.W., Jr.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef]
- Anand, S.P.; Finzi, A. Understudied Factors Influencing Fc-Mediated Immune Responses against Viral Infections. Vaccines 2019, 7, 103. [Google Scholar] [CrossRef]
- Bruhns, P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood 2012, 119, 5640–5649. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Kinet, J.P. Fc receptors. Annu. Rev. Immunol. 1991, 9, 457–492. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Bruhns, P.; Horiuchi, K.; Ravetch, J.V. FcgammaRIV: A novel FcR with distinct IgG subclass specificity. Immunity 2005, 23, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.; DiLillo, D.J.; Bournazos, S.; Li, F.; Ravetch, J.V. Mouse model recapitulating human Fcgamma receptor structural and functional diversity. Proc. Natl. Acad. Sci. USA 2012, 109, 6181–6186. [Google Scholar] [CrossRef] [PubMed]
- Irani, V.; Guy, A.J.; Andrew, D.; Beeson, J.G.; Ramsland, P.A.; Richards, J.S. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol. Immunol. 2015, 67, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Rivkin, A.; Pham, T. Panitumumab: Human monoclonal antibody against epidermal growth factor receptors for the treatment of metastatic colorectal cancer. Clin. Ther. 2008, 30, 14–30. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef]
- An, Z.; Forrest, G.; Moore, R.; Cukan, M.; Haytko, P.; Huang, L.; Vitelli, S.; Zhao, J.Z.; Lu, P.; Hua, J.; et al. IgG2m4, an engineered antibody isotype with reduced Fc function. MAbs 2009, 1, 572–579. [Google Scholar] [CrossRef]
- Pincetic, A.; Bournazos, S.; DiLillo, D.J.; Maamary, J.; Wang, T.T.; Dahan, R.; Fiebiger, B.M.; Ravetch, J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014, 15, 707–716. [Google Scholar] [CrossRef]
- Bondt, A.; Rombouts, Y.; Selman, M.H.; Hensbergen, P.J.; Reiding, K.R.; Hazes, J.M.; Dolhain, R.J.; Wuhrer, M. Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol. Cell Proteom. 2014, 13, 3029–3039. [Google Scholar] [CrossRef] [PubMed]
- Yamada, E.; Tsukamoto, Y.; Sasaki, R.; Yagyu, K.; Takahashi, N. Structural changes of immunoglobulin G oligosaccharides with age in healthy human serum. Glycoconj. J. 1997, 14, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.S.; Wu, X.; Kulhavy, R.; Tomana, M.; Novak, J.; Moldoveanu, Z.; Brown, R.; Goepfert, P.A.; Mestecky, J. Increased levels of galactose-deficient IgG in sera of HIV-1-infected individuals. AIDS 2005, 19, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef]
- Kellner, C.; Otte, A.; Cappuzzello, E.; Klausz, K.; Peipp, M. Modulating Cytotoxic Effector Functions by Fc Engineering to Improve Cancer Therapy. Transfus. Med. Hemother. 2017, 44, 327–336. [Google Scholar] [CrossRef]
- Monnet, C.; Jorieux, S.; Souyris, N.; Zaki, O.; Jacquet, A.; Fournier, N.; Crozet, F.; de Romeuf, C.; Bouayadi, K.; Urbain, R.; et al. Combined glyco- and protein-Fc engineering simultaneously enhance cytotoxicity and half-life of a therapeutic antibody. MAbs 2014, 6, 422–436. [Google Scholar] [CrossRef]
- Gomez-Roman, V.R.; Florese, R.H.; Peng, B.; Montefiori, D.C.; Kalyanaraman, V.S.; Venzon, D.; Srivastava, I.; Barnett, S.W.; Robert-Guroff, M. An adenovirus-based HIV subtype B prime/boost vaccine regimen elicits antibodies mediating broad antibody-dependent cellular cytotoxicity against non-subtype B HIV strains. J. Acquir. Immune Defic. Syndr. 2006, 43, 270–277. [Google Scholar] [CrossRef]
- Richard, J.; Prevost, J.; Alsahafi, N.; Ding, S.; Finzi, A. Impact of HIV-1 Envelope Conformation on ADCC Responses. Trends Microbiol. 2018, 26, 253–265. [Google Scholar] [CrossRef]
- Saphire, E.O.; Schendel, S.L.; Fusco, M.L.; Gangavarapu, K.; Gunn, B.M.; Wec, A.Z.; Halfmann, P.J.; Brannan, J.M.; Herbert, A.S.; Qiu, X.; et al. Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 2018, 174, 938–952. [Google Scholar] [CrossRef]
- Garcia, G.; Arango, M.; Perez, A.B.; Fonte, L.; Sierra, B.; Rodriguez-Roche, R.; Aguirre, E.; Fiterre, I.; Guzman, M.G. Antibodies from patients with dengue viral infection mediate cellular cytotoxicity. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2006, 37, 53–57. [Google Scholar] [CrossRef]
- Zhang, M.; Daniel, S.; Huang, Y.; Chancey, C.; Huang, Q.; Lei, Y.F.; Grinev, A.; Mostowski, H.; Rios, M.; Dayton, A. Anti-West Nile virus activity of in vitro expanded human primary natural killer cells. BMC Immunol. 2010, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.C.; O’Connell, R.J. Progress in HIV vaccine development. Hum. Vaccines Immunother. 2017, 13, 1018–1030. [Google Scholar] [CrossRef]
- Lee, W.S.; Kent, S.J. Anti-HIV-1 antibody-dependent cellular cytotoxicity: Is there more to antibodies than neutralization? Curr. Opin. HIV AIDS 2018, 13, 160–166. [Google Scholar] [CrossRef]
- Bruel, T.; Guivel-Benhassine, F.; Lorin, V.; Lortat-Jacob, H.; Baleux, F.; Bourdic, K.; Noel, N.; Lambotte, O.; Mouquet, H.; Schwartz, O. Lack of ADCC Breadth of Human Nonneutralizing Anti-HIV-1 Antibodies. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Chung, A.W.; Ghebremichael, M.; Robinson, H.; Brown, E.; Choi, I.; Lane, S.; Dugast, A.S.; Schoen, M.K.; Rolland, M.; Suscovich, T.J.; et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci. Transl. Med. 2014, 6, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, N.; Chester, C.; Yonezawa, A.; Zhao, X.; Kohrt, H.E. Enhancement of antibody-dependent cell mediated cytotoxicity: A new era in cancer treatment. Immuno Targets Ther. 2015, 4, 91–100. [Google Scholar] [CrossRef]
- Kramski, M.; Schorcht, A.; Johnston, A.P.; Lichtfuss, G.F.; Jegaskanda, S.; De Rose, R.; Stratov, I.; Kelleher, A.D.; French, M.A.; Center, R.J.; et al. Role of monocytes in mediating HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. Methods 2012, 384, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Mullarkey, C.E.; Bailey, M.J.; Golubeva, D.A.; Tan, G.S.; Nachbagauer, R.; He, W.; Novakowski, K.E.; Bowdish, D.M.; Miller, M.S.; Palese, P. Broadly Neutralizing Hemagglutinin Stalk-Specific Antibodies Induce Potent Phagocytosis of Immune Complexes by Neutrophils in an Fc-Dependent Manner. MBio 2016, 7. [Google Scholar] [CrossRef]
- Barker, E.; Reisfeld, R.A. A mechanism for neutrophil-mediated lysis of human neuroblastoma cells. Cancer Res. 1993, 53, 362–367. [Google Scholar]
- Ackerman, M.E.; Moldt, B.; Wyatt, R.T.; Dugast, A.S.; McAndrew, E.; Tsoukas, S.; Jost, S.; Berger, C.T.; Sciaranghella, G.; Liu, Q.; et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 2011, 366, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Horner, H.; Frank, C.; Dechant, C.; Repp, R.; Glennie, M.; Herrmann, M.; Stockmeyer, B. Intimate cell conjugate formation and exchange of membrane lipids precede apoptosis induction in target cells during antibody-dependent, granulocyte-mediated cytotoxicity. J. Immunol. 2007, 179, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Moeller, E. Contact-Induced Cytotoxicity by Lymphoid Cells Containing Foreign Isoantigens. Science 1965, 147, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.K.; Ackerman, M.E.; Scarlatti, G.; Moog, C.; Robert-Guroff, M.; Kent, S.J.; Overbaugh, J.; Reeves, R.K.; Ferrari, G.; Thyagarajan, B. Knowns and Unknowns of Assaying Antibody-Dependent Cell-Mediated Cytotoxicity Against HIV-1. Front. Immunol. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Tyler, D.S.; Stanley, S.D.; Nastala, C.A.; Austin, A.A.; Bartlett, J.A.; Stine, K.C.; Lyerly, H.K.; Bolognesi, D.P.; Weinhold, K.J. Alterations in antibody-dependent cellular cytotoxicity during the course of HIV-1 infection. Humoral and cellular defects. J. Immunol. 1990, 144, 3375–3384. [Google Scholar]
- Baum, L.L.; Cassutt, K.J.; Knigge, K.; Khattri, R.; Margolick, J.; Rinaldo, C.; Kleeberger, C.A.; Nishanian, P.; Henrard, D.R.; Phair, J. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1996, 157, 2168–2173. [Google Scholar]
- Kurane, I.; Hebblewaite, D.; Brandt, W.E.; Ennis, F.A. Lysis of dengue virus-infected cells by natural cell-mediated cytotoxicity and antibody-dependent cell-mediated cytotoxicity. J. Virol. 1984, 52, 223–230. [Google Scholar] [CrossRef]
- Srivastava, V.; Yang, Z.; Hung, I.F.; Xu, J.; Zheng, B.; Zhang, M.Y. Identification of dominant antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin antigen of pandemic H1N1 influenza virus. J. Virol. 2013, 87, 5831–5840. [Google Scholar] [CrossRef][Green Version]
- Ye, Z.W.; Yuan, S.; Poon, K.M.; Wen, L.; Yang, D.; Sun, Z.; Li, C.; Hu, M.; Shuai, H.; Zhou, J.; et al. Antibody-Dependent Cell-Mediated Cytotoxicity Epitopes on the Hemagglutinin Head Region of Pandemic H1N1 Influenza Virus Play Detrimental Roles in H1N1-Infected Mice. Front. Immunol. 2017, 8, 317. [Google Scholar] [CrossRef]
- Radosevic, K.; Garritsen, H.S.; Van Graft, M.; De Grooth, B.G.; Greve, J. A simple and sensitive flow cytometric assay for the determination of the cytotoxic activity of human natural killer cells. J. Immunol. Methods 1990, 135, 81–89. [Google Scholar] [CrossRef][Green Version]
- de Vries, R.D.; Nieuwkoop, N.J.; van der Klis, F.R.M.; Koopmans, M.P.G.; Krammer, F.; Rimmelzwaan, G.F. Primary Human Influenza B Virus Infection Induces Cross-Lineage Hemagglutinin Stalk-Specific Antibodies Mediating Antibody-Dependent Cellular Cytoxicity. J. Infect. Dis. 2017, 217, 3–11. [Google Scholar] [CrossRef]
- Chai, N.; Swem, L.R.; Park, S.; Nakamura, G.; Chiang, N.; Estevez, A.; Fong, R.; Kamen, L.; Kho, E.; Reichelt, M.; et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat. Commun. 2017, 8, 14234. [Google Scholar] [CrossRef]
- Alter, G.; Malenfant, J.M.; Altfeld, M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 2004, 294, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Krzewski, K.; Gil-Krzewska, A.; Nguyen, V.; Peruzzi, G.; Coligan, J.E. LAMP1/CD107a is required for efficient perforin delivery to lytic granules and NK-cell cytotoxicity. Blood 2013, 121, 4672–4683. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.W.; Navis, M.; Isitman, G.; Wren, L.; Silvers, J.; Amin, J.; Kent, S.J.; Stratov, I. Activation of NK cells by ADCC antibodies and HIV disease progression. J. Acquir. Immune Defic. Syndr. 2011, 58, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Madhavi, V.; Navis, M.; Chung, A.W.; Isitman, G.; Wren, L.H.; De Rose, R.; Kent, S.J.; Stratov, I. Activation of NK cells by HIV-specific ADCC antibodies: Role for granulocytes in expressing HIV-1 peptide epitopes. Hum. Vaccines Immunother. 2013, 9, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.J.; Garvin, D.; Paguio, A.; Moravec, R.; Engel, L.; Fan, F.; Surowy, T. Development of a robust reporter-based ADCC assay with frozen, thaw-and-use cells to measure Fc effector function of therapeutic antibodies. J. Immunol. Methods 2014, 414, 69–81. [Google Scholar] [CrossRef]
- Tan, G.S.; Leon, P.E.; Albrecht, R.A.; Margine, I.; Hirsh, A.; Bahl, J.; Krammer, F. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLoS Pathog. 2016, 12, e1005578. [Google Scholar] [CrossRef]
- Amanat, F.; Meade, P.; Strohmeier, S.; Krammer, F. Cross-reactive antibodies binding to H4 hemagglutinin protect against a lethal H4N6 influenza virus challenge in the mouse model. Emerg. Microbes Infect. 2019, 8, 155–168. [Google Scholar] [CrossRef]
- Cox, F.; Kwaks, T.; Brandenburg, B.; Koldijk, M.H.; Klaren, V.; Smal, B.; Korse, H.J.; Geelen, E.; Tettero, L.; Zuijdgeest, D.; et al. HA Antibody-Mediated FcgammaRIIIa Activity Is Both Dependent on FcR Engagement and Interactions between HA and Sialic Acids. Front. Immunol. 2016, 7, 399. [Google Scholar] [CrossRef]
- He, W.; Tan, G.S.; Mullarkey, C.E.; Lee, A.J.; Lam, M.M.; Krammer, F.; Henry, C.; Wilson, P.C.; Ashkar, A.A.; Palese, P.; et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl. Acad. Sci. USA 2016, 113, 11931–11936. [Google Scholar] [CrossRef] [PubMed]
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.F.; Huang, Y.; Kaur, K.; Popova, L.I.; Ho, I.Y.; Pauli, N.T.; Henry Dunand, C.J.; Taylor, W.M.; Lim, S.; Huang, M.; et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015, 7, 316ra192. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef]
- Tan, G.S.; Krammer, F.; Eggink, D.; Kongchanagul, A.; Moran, T.M.; Palese, P. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J. Virol. 2012, 86, 6179–6188. [Google Scholar] [CrossRef]
- Desbien, A.L.; Van Hoeven, N.; Reed, S.J.; Casey, A.C.; Laurance, J.D.; Baldwin, S.L.; Duthie, M.S.; Reed, S.G.; Carter, D. Development of a high density hemagglutinin protein microarray to determine the breadth of influenza antibody responses. Biotechniques 2013, 54, 345–348. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Petersen, E.; Moran, T.M.; Palese, P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010, 6, e1000796. [Google Scholar] [CrossRef]
- Yasugi, M.; Kubota-Koketsu, R.; Yamashita, A.; Kawashita, N.; Du, A.; Sasaki, T.; Nishimura, M.; Misaki, R.; Kuhara, M.; Boonsathorn, N.; et al. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog. 2013, 9, e1003150. [Google Scholar] [CrossRef]
- Lang, S.; Xie, J.; Zhu, X.; Wu, N.C.; Lerner, R.A.; Wilson, I.A. Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in VH1-69 Antibody Orientation on the HA Stem. Cell Rep. 2017, 20, 2935–2943. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Laursen, N.S.; Wilson, I.A. Broadly neutralizing antibodies against influenza viruses. Antivir. Res. 2013, 98, 476–483. [Google Scholar] [CrossRef]
- Krammer, F. Novel universal influenza virus vaccine approaches. Curr. Opin. Virol. 2016, 17, 95–103. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1, 1–10. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Krammer, F. Universal influenza virus vaccines and therapeutic antibodies. Clin. Microbiol. Infect. 2017, 23, 222–228. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef]
- Brandenburg, B.; Koudstaal, W.; Goudsmit, J.; Klaren, V.; Tang, C.; Bujny, M.V.; Korse, H.J.; Kwaks, T.; Otterstrom, J.J.; Juraszek, J.; et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS ONE 2013, 8, e80034. [Google Scholar] [CrossRef]
- Bangaru, S.; Zhang, H.; Gilchuk, I.M.; Voss, T.G.; Irving, R.P.; Gilchuk, P.; Matta, P.; Zhu, X.; Lang, S.; Nieusma, T.; et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018, 9, 2669. [Google Scholar] [CrossRef]
- Watanabe, A.; McCarthy, K.R.; Kuraoka, M.; Schmidt, A.G.; Adachi, Y.; Onodera, T.; Tonouchi, K.; Caradonna, T.M.; Bajic, G.; Song, S.; et al. Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 2019, 177, 1124–1135. [Google Scholar] [CrossRef]
- Bangaru, S.; Lang, S.; Schotsaert, M.; Vanderven, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 2019, 177, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Leon, P.E.; He, W.; Mullarkey, C.E.; Bailey, M.J.; Miller, M.S.; Krammer, F.; Palese, P.; Tan, G.S. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc. Natl. Acad. Sci. USA 2016, 113, E5944–E5951. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Chen, J.; Li, R.; Zhang, M.; Wang, G.; Stegalkina, S.; Zhang, L.; Chen, J.; Cao, J.; Bi, X.; et al. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, A.; Pleschka, S. Influenza H3N2 Vaccines: Recent Challenges. Trends Microbiol. 2018, 26, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Neirynck, S.; Deroo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef]
- Schotsaert, M.; De Filette, M.; Fiers, W.; Saelens, X. Universal M2 ectodomain-based influenza A vaccines: Preclinical and clinical developments. Expert Rev. Vaccines 2009, 8, 499–508. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Monto, A.S. Neuraminidase, the Forgotten Surface Antigen, Emerges as an Influenza Vaccine Target for Broadened Protection. J. Infect. Dis. 2019, 219, S75–S80. [Google Scholar] [CrossRef]
- Wohlbold, T.J.; Nachbagauer, R.; Xu, H.; Tan, G.S.; Hirsh, A.; Brokstad, K.A.; Cox, R.J.; Palese, P.; Krammer, F. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. MBio 2015, 6, e02556. [Google Scholar] [CrossRef]
- Wohlbold, T.J.; Krammer, F. In the shadow of hemagglutinin: A growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 2014, 6, 2465–2494. [Google Scholar] [CrossRef]
- Stadlbauer, D.; Zhu, X.; McMahon, M.; Turner, J.S.; Wohlbold, T.J.; Schmitz, A.J.; Strohmeier, S.; Yu, W.; Nachbagauer, R.; Mudd, P.A.; et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 2019, 366, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Cady, S.D.; Luo, W.; Hu, F.; Hong, M. Structure and function of the influenza A M2 proton channel. Biochemistry 2009, 48, 7356–7364. [Google Scholar] [CrossRef] [PubMed]
- Pielak, R.M.; Chou, J.J. Influenza M2 proton channels. Biochim. Biophys. Acta 2011, 1808, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhou, L.; Chen, Y.H. Immunization with high epitope density of M2e derived from 2009 pandemic H1N1 elicits protective immunity in mice. Vaccine 2012, 30, 3463–3469. [Google Scholar] [CrossRef] [PubMed]
- El Bakkouri, K.; Descamps, F.; De Filette, M.; Smet, A.; Festjens, E.; Birkett, A.; Van Rooijen, N.; Verbeek, S.; Fiers, W.; Saelens, X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011, 186, 1022–1031. [Google Scholar] [CrossRef]
- Lee, Y.T.; Kim, K.H.; Ko, E.J.; Lee, Y.N.; Kim, M.C.; Kwon, Y.M.; Tang, Y.; Cho, M.K.; Lee, Y.J.; Kang, S.M. New vaccines against influenza virus. Clin. Exp. Vaccine Res. 2014, 3, 12–28. [Google Scholar] [CrossRef]
- Schepens, B.; De Vlieger, D.; Saelens, X. Vaccine options for influenza: Thinking small. Curr. Opin. Immunol. 2018, 53, 22–29. [Google Scholar] [CrossRef]
- Lee, Y.N.; Lee, Y.T.; Kim, M.C.; Hwang, H.S.; Lee, J.S.; Kim, K.H.; Kang, S.M. Fc receptor is not required for inducing antibodies but plays a critical role in conferring protection after influenza M2 vaccination. Immunology 2014, 143, 300–309. [Google Scholar] [CrossRef]
- Kolpe, A.; Schepens, B.; Fiers, W.; Saelens, X. M2-based influenza vaccines: Recent advances and clinical potential. Expert Rev. Vaccines 2017, 16, 123–136. [Google Scholar] [CrossRef]
- Song, A.; Myojo, K.; Laudenslager, J.; Harada, D.; Miura, T.; Suzuki, K.; Kuni-Kamochi, R.; Soloff, R.; Ohgami, K.; Kanda, Y. Evaluation of a fully human monoclonal antibody against multiple influenza A viral strains in mice and a pandemic H1N1 strain in nonhuman primates. Antivir. Res. 2014, 111, 60–68. [Google Scholar] [CrossRef]
- Virelizier, J.L.; Allison, A.C.; Oxford, J.S.; Schild, G.C. Early presence of ribonucleoprotein antigen on surface of influenza virus-infected cells. Nature 1977, 266, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Frank, E.; Gerhard, W. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J. Immunol. 1981, 126, 1814–1819. [Google Scholar]
- Antrobus, R.D.; Lillie, P.J.; Berthoud, T.K.; Spencer, A.J.; McLaren, J.E.; Ladell, K.; Lambe, T.; Milicic, A.; Price, D.A.; Hill, A.V.; et al. A T cell-inducing influenza vaccine for the elderly: Safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS ONE 2012, 7, e48322. [Google Scholar] [CrossRef]
- Zheng, M.; Luo, J.; Chen, Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection 2014, 42, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, T.K.; Hamill, M.; Lillie, P.J.; Hwenda, L.; Collins, K.A.; Ewer, K.J.; Milicic, A.; Poyntz, H.C.; Lambe, T.; Fletcher, H.A.; et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin. Infect. Dis. 2011, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Co, M.D.T.; Cruz, J.; Subbarao, K.; Ennis, F.A.; Terajima, M. Induction of H7N9-Cross-Reactive Antibody-Dependent Cellular Cytotoxicity Antibodies by Human Seasonal Influenza A Viruses that are Directed Toward the Nucleoprotein. J. Infect. Dis. 2017, 215, 818–823. [Google Scholar] [CrossRef]
- Carragher, D.M.; Kaminski, D.A.; Moquin, A.; Hartson, L.; Randall, T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J. Immunol. 2008, 181, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Florek, N.W.; Weinfurter, J.T.; Jegaskanda, S.; Brewoo, J.N.; Powell, T.D.; Young, G.R.; Das, S.C.; Hatta, M.; Broman, K.W.; Hungnes, O.; et al. Modified vaccinia virus Ankara encoding influenza virus hemagglutinin induces heterosubtypic immunity in macaques. J. Virol. 2014, 88, 13418–13428. [Google Scholar] [CrossRef]
- Vanderven, H.A.; Liu, L.; Ana-Sosa-Batiz, F.; Nguyen, T.H.; Wan, Y.; Wines, B.; Hogarth, P.M.; Tilmanis, D.; Reynaldi, A.; Parsons, M.S.; et al. Fc functional antibodies in humans with severe H7N9 and seasonal influenza. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Horns, F.; Vollmers, C.; Croote, D.; Mackey, S.F.; Swan, G.E.; Dekker, C.L.; Davis, M.M.; Quake, S.R. Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. Elife 2016, 5, e16578. [Google Scholar] [CrossRef]
- Wang, J.; Liu, M.; Ding, N.; Li, Y.; Shao, J.; Zhu, M.; Xie, Z.; Sun, K. Vaccine based on antibody-dependent cell-mediated cytotoxicity epitope on the H1N1 influenza virus increases mortality in vaccinated mice. Biochem. Biophys. Res. Commun. 2018, 503, 1874–1879. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Okayama, H.; Ashizawa, M.; Noda, M.; Aoto, K.; Saito, M.; Monma, T.; Ohki, S.; Shibata, M.; Takenoshita, S.; et al. Augmentation of antibody-dependent cellular cytotoxicity with defucosylated monoclonal antibodies in patients with GI-tract cancer. Oncol. Lett. 2018, 15, 2604–2610. [Google Scholar] [CrossRef] [PubMed]
- Kohrt, H.E.; Houot, R.; Marabelle, A.; Cho, H.J.; Osman, K.; Goldstein, M.; Levy, R.; Brody, J. Combination strategies to enhance antitumor ADCC. Immunotherapy 2012, 4, 511–527. [Google Scholar] [CrossRef] [PubMed]
- Lazar, G.A.; Dang, W.; Karki, S.; Vafa, O.; Peng, J.S.; Hyun, L.; Chan, C.; Chung, H.S.; Eivazi, A.; Yoder, S.C.; et al. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. USA 2006, 103, 4005–4010. [Google Scholar] [CrossRef]
- Ming Lim, C.; Stephenson, R.; Salazar, A.M.; Ferris, R.L. TLR3 agonists improve the immunostimulatory potential of cetuximab against EGFR(+) head and neck cancer cells. Oncoimmunology 2013, 2, e24677. [Google Scholar] [CrossRef]
- Roda, J.M.; Parihar, R.; Carson, W.E., 3rd. CpG-containing oligodeoxynucleotides act through TLR9 to enhance the NK cell cytokine response to antibody-coated tumor cells. J. Immunol. 2005, 175, 1619–1627. [Google Scholar] [CrossRef]
- Chung, A.W.; Isitman, G.; Navis, M.; Kramski, M.; Center, R.J.; Kent, S.J.; Stratov, I. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc. Natl. Acad. Sci. USA 2011, 108, 7505–7510. [Google Scholar] [CrossRef]
- Smalls-Mantey, A.; Doria-Rose, N.; Klein, R.; Patamawenu, A.; Migueles, S.A.; Ko, S.Y.; Hallahan, C.W.; Wong, H.; Liu, B.; You, L.; et al. Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. J. Virol. 2012, 86, 8672–8680. [Google Scholar] [CrossRef]
- Boge, M.; Wyss, S.; Bonifacino, J.S.; Thali, M. Membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 1998, 273, 15773–15778. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Schmitz, N.; Storni, T.; Bachmann, M.F. Influenza A vaccine based on the extracellular domain of M2: Weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004, 172, 5598–5605. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S. The Potential Role of Fc-Receptor Functions in the Development of a Universal Influenza Vaccine. Vaccines 2018, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Kosik, I.; Angeletti, D.; Gibbs, J.S.; Angel, M.; Takeda, K.; Kosikova, M.; Nair, V.; Hickman, H.D.; Xie, H.; Brooke, C.B.; et al. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J. Exp. Med. 2019, 216, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Mohan, T.; Chang, T.Z.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.M.; Kang, S.M.; Compans, R.W.; Champion, J.A.; Wang, B.Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Sheng, Z.; Sreenivasan, C.C.; Wang, D.; Li, F. Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function. Viruses 2020, 12, 276. https://doi.org/10.3390/v12030276
Gao R, Sheng Z, Sreenivasan CC, Wang D, Li F. Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function. Viruses. 2020; 12(3):276. https://doi.org/10.3390/v12030276
Chicago/Turabian StyleGao, Rongyuan, Zizhang Sheng, Chithra C. Sreenivasan, Dan Wang, and Feng Li. 2020. "Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function" Viruses 12, no. 3: 276. https://doi.org/10.3390/v12030276
APA StyleGao, R., Sheng, Z., Sreenivasan, C. C., Wang, D., & Li, F. (2020). Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function. Viruses, 12(3), 276. https://doi.org/10.3390/v12030276





