Superinfection by PHYVV Alters the Recovery Process in PepGMV-Infected Pepper Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Viral Clones
2.3. Virus Inoculation
2.4. Superinfection Assays
2.5. Viral DNA Accumulation in Infected Plants
2.6. Isolation of PepGMV Minichromosomes
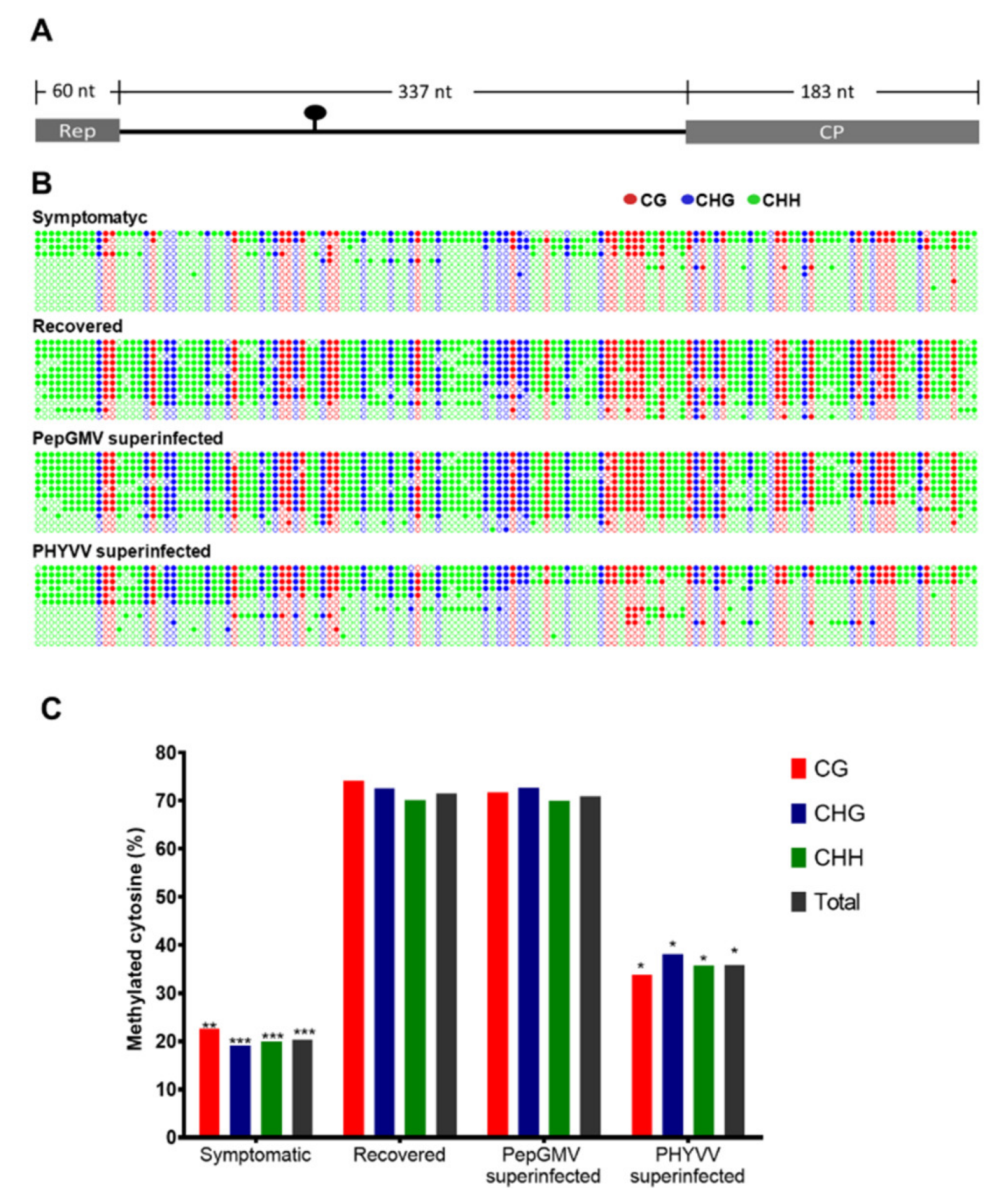
2.7. Methylation Density Analysis
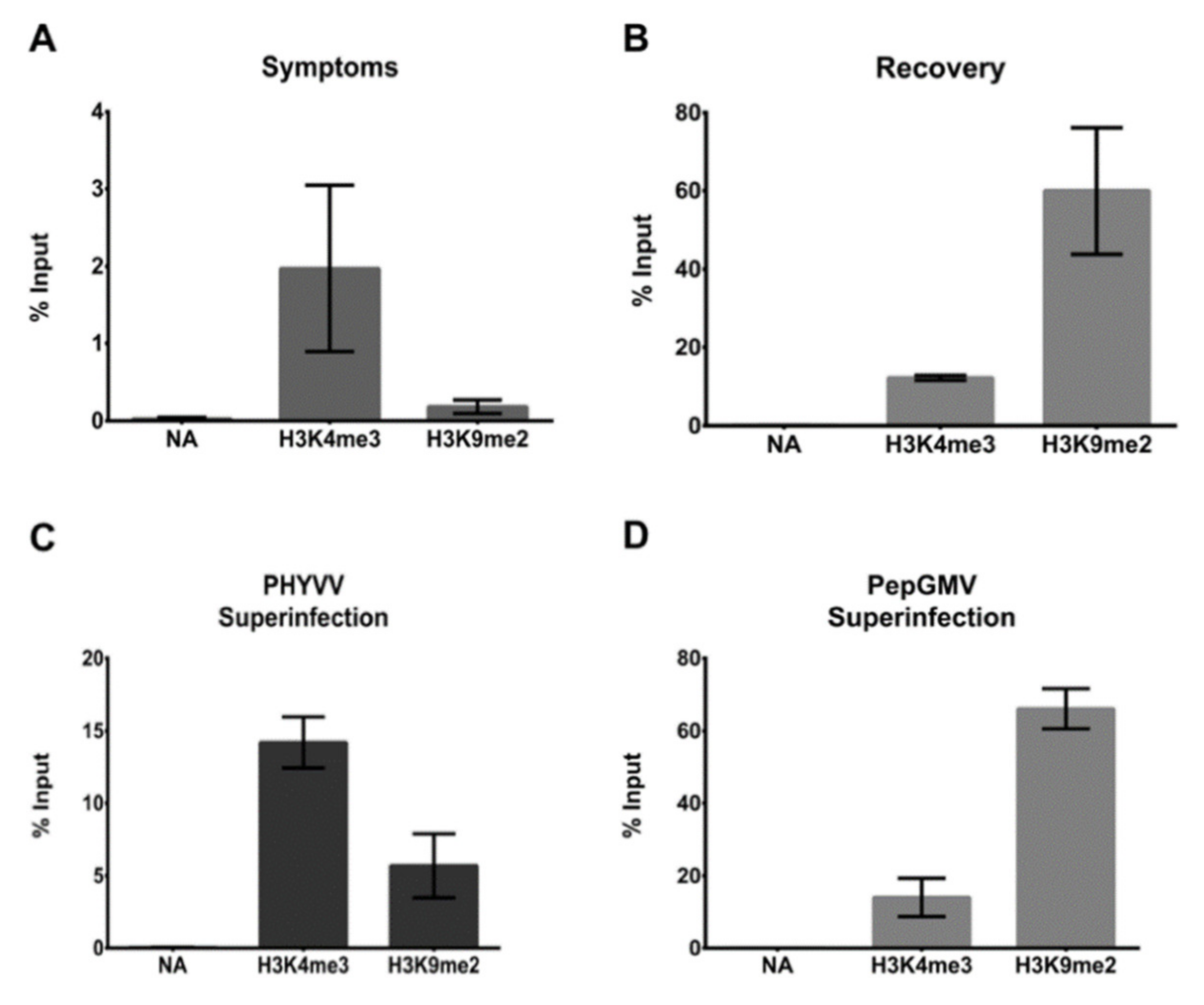
2.8. ChIP-qPCR Analysis
2.9. PHYVV Gene Expression Analysis
2.10. Suppressor Silencing Analysis
2.11. GFP Expression Analysis
2.12. Chop-PCR Analysis
3. Results
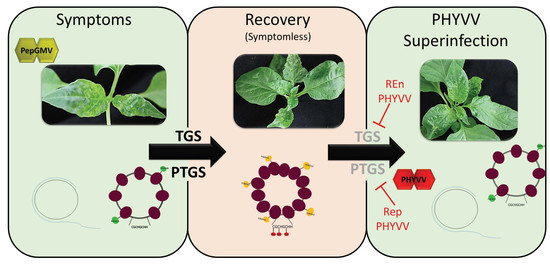
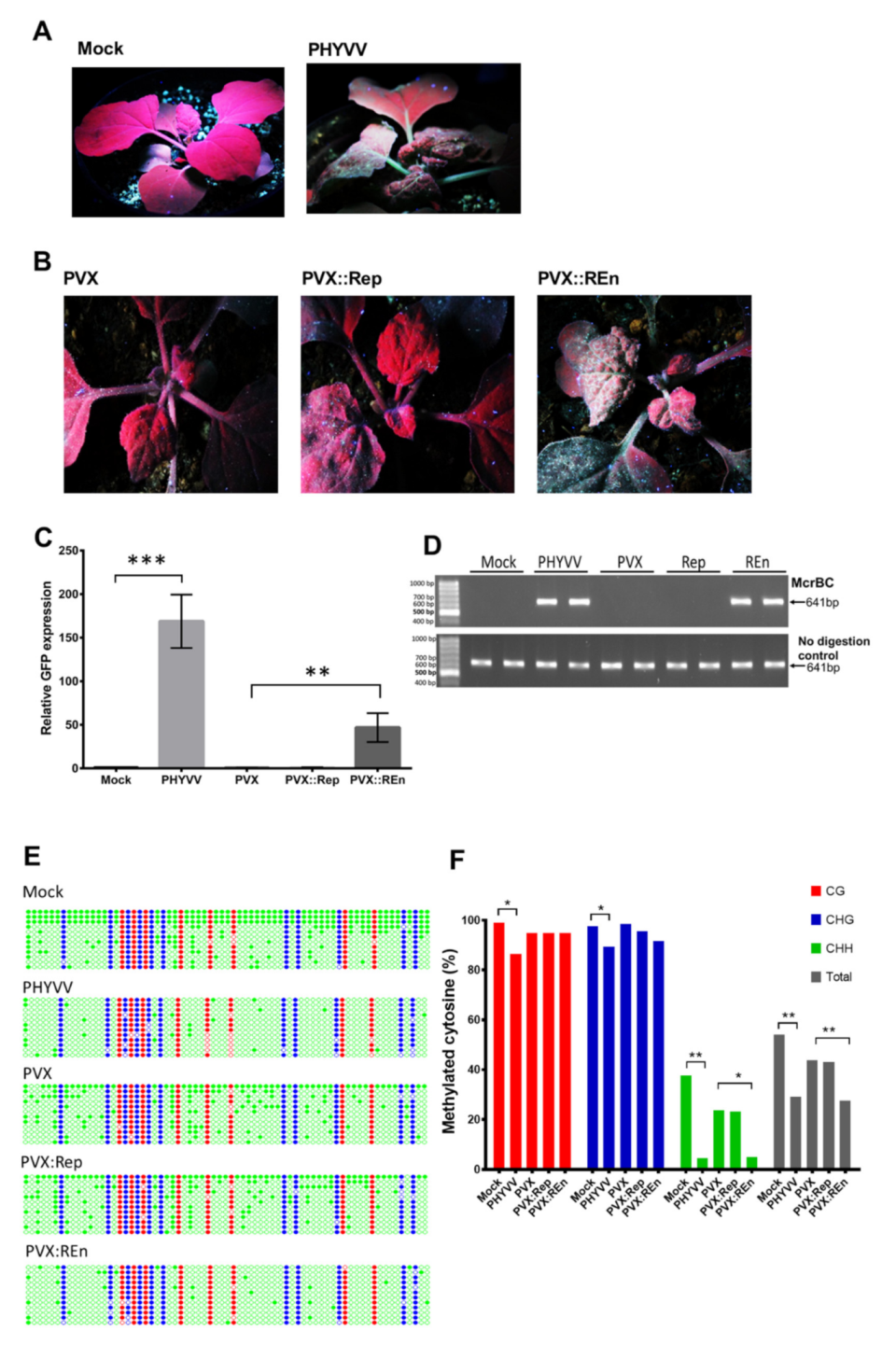
3.1. PHYVV Superinfection Reverts Host Recovery in PepGMV-Infected Pepper Plants
3.2. DNA Methylation Level of PepGMV Genome is Reduced in PHYVV Superinfected Plants
3.3. CHIP-qPCR Analysis
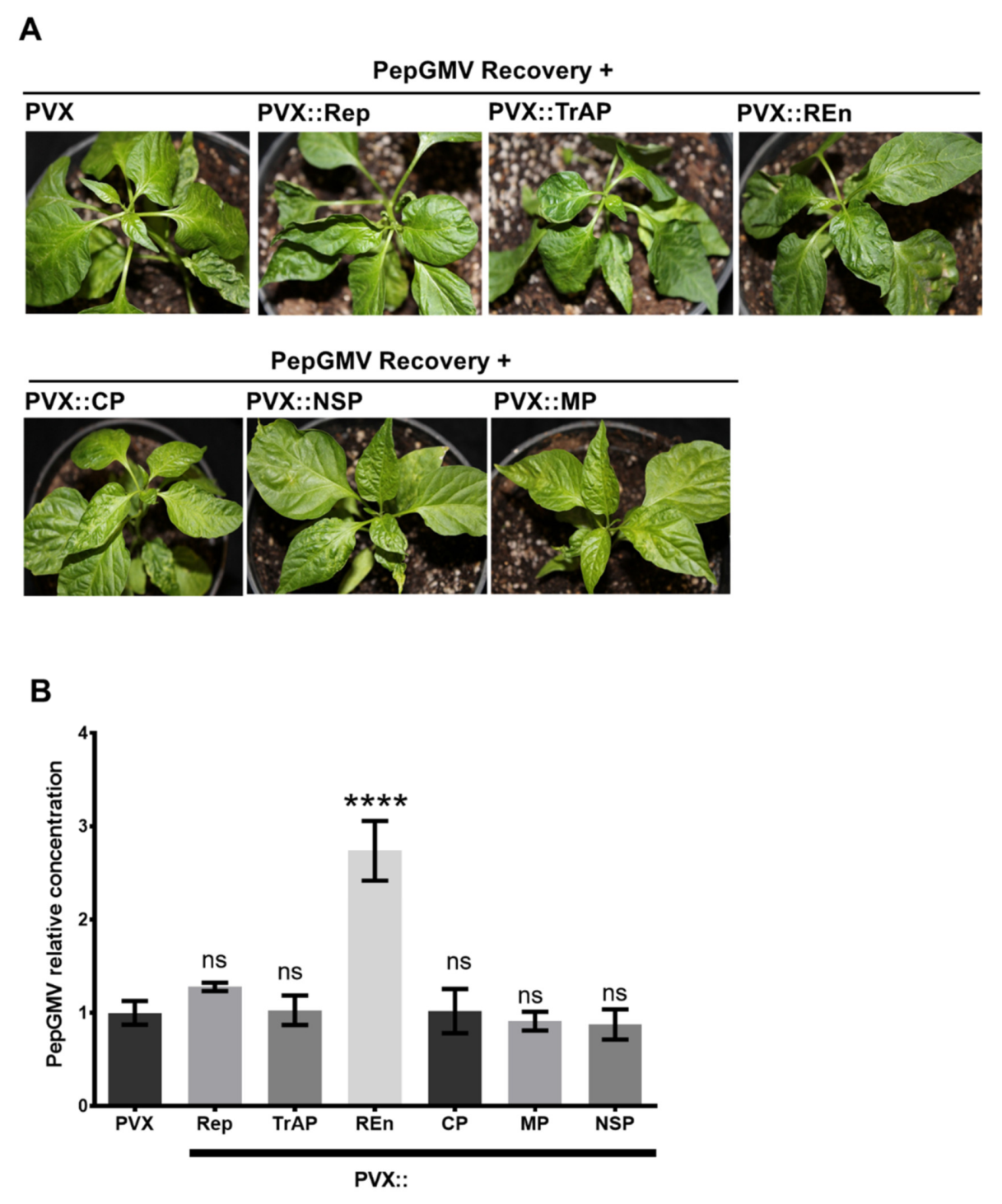
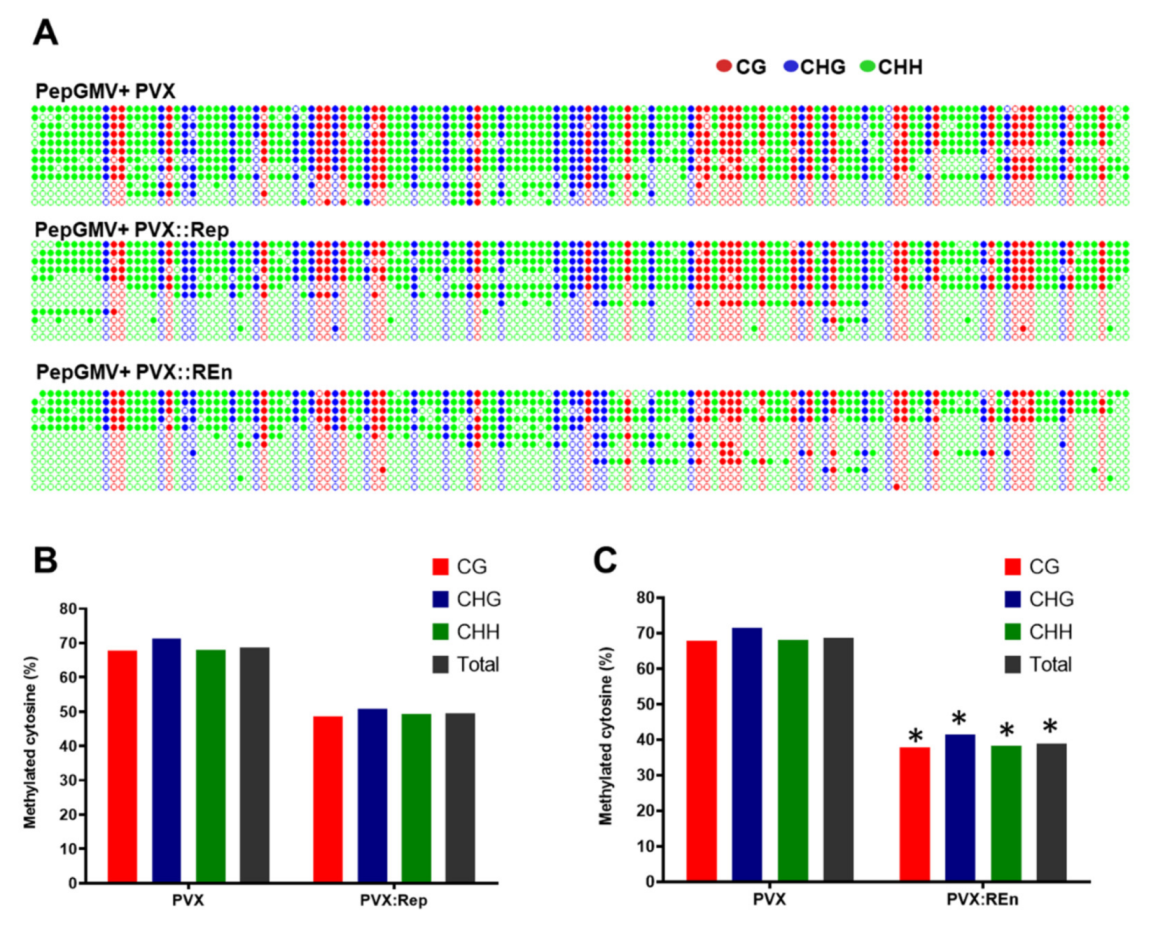
3.4. PepGMV Accumulation in Recovery Plants is Enhanced by PHYVV Encoded Rep and REn Proteins
3.5. Expression of PHYVV REn Reduces the Levels of PepGMV DNA Methylation
3.6. PHYVV REn Is a TGS Silencing Suppressor
3.7. PHYVV Rep Is a PTGS Silencing Suppressor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 2 July 2019).
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.F.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Tripp, J.; Lozoya-Gloria, E.; Rivera-Bustamante, R.F. Symptom remission and specific resistance of pepper plants after infection by Pepper golden mosaic virus. Phytopathology 2007, 97, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Méndez-Lozano, J.; Torres-Pacheco, I.; Fauquet, C.M.; Rivera-Bustamante, R.F. Interactions Between Geminiviruses in a Naturally Occurring Mixture: Pepper huasteco virus and Pepper golden mosaic virus. Phytopathology 2003, 93, 270–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rentería-Canett, I.; Xoconostle-Cázares, B.; Ruiz-Medrano, R.; Rivera-Bustamante, R.F. Geminivirus mixed infection on pepper plants: Synergistic interaction between PHYVV and PepGMV. Virol. J. 2011, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Rodelo-Urrego, M.; Pagán, I.; González-Jara, P.; Betancourt, M.; Moreno-Letelier, A.; Ayllõn, M.A.; Fraile, A.; Piñero, D.; García-Arenal, F. Landscape heterogeneity shapes host-parasite interactions and results in apparent plant-virus codivergence. Mol. Ecol. 2013, 22, 2325–2340. [Google Scholar] [CrossRef]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Voinnet, O.; Pinto, Y.M.; Baulcombe, D.C. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 1999, 96, 14147–14152. [Google Scholar] [CrossRef] [Green Version]
- Raja, P.; Wolf, J.N.; Bisaro, D.M. RNA silencing directed against geminiviruses: Post-transcriptional and epigenetic components. Biochim. Biophys. Acta Gene Regul. Mech. 2010, 1799, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Pita, J.; Fauquet, C.M. Short Interfering RNA Accumulation Correlates with Host Recovery in DNA Virus-Infected Hosts, and Gene Silencing Targets Specific Viral Sequences. J. Virol. 2006, 80, 1064. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Negrete, E.A.; Carrillo-Tripp, J.; Rivera-Bustamante, R.F. RNA Silencing against Geminivirus: Complementary Action of Posttranscriptional Gene Silencing and Transcriptional Gene Silencing in Host Recovery. J. Virol. 2009, 83, 1332–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramesh, S.V.; Sahu, P.P.; Prasad, M.; Praveen, S.; Pappu, H.R. Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race. Viruses 2017, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abouzid, A.M.; Frischmuth, T.; Jeske, H. A putative replicative form of the abutilon mosaic virus (gemini group) in a chromatin-like structure. MGG Mol. Gen. Genet. 1988, 212, 252–258. [Google Scholar] [CrossRef]
- Pilartz, M.; Jeske, H. Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 1992, 189, 800–802. [Google Scholar] [CrossRef]
- Ceniceros-Ojeda, E.A.; Rodríguez-Negrete, E.A.; Rivera-Bustamante, R.F. Two Populations of Viral Minichromosomes Are Present in a Geminivirus-Infected Plant Showing Symptom Remission (Recovery). J. Virol. 2016, 90, 3828–3838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [Green Version]
- Bisaro, D.M. Silencing suppression by geminivirus proteins. Virology 2006, 344, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Csorba, T.; Kontra, L.; Burgyán, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Rishishwar, R.; Dasgupta, I. Suppressors of RNA silencing encoded by geminiviruses and associated DNA satellites. Virus Dis. 2019, 30, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Dang, M.; Hou, H.; Mei, Y.; Qian, Y.; Zhou, X. Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 2014, 95, 2082–2088. [Google Scholar] [CrossRef] [PubMed]
- Vinutha, T.; Kumar, G.; Garg, V.; Canto, T.; Palukaitis, P.; Ramesh, S.V.; Praveen, S. Tomato geminivirus encoded RNAi suppressor protein, AC4 interacts with host AGO4 and precludes viral DNA methylation. Gene 2018, 678, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, R.C.; Asad, S.; Wolf, J.N.; Mohannath, G.; Bisaro, D.M. Geminivirus AL2 and L2 Proteins Suppress Transcriptional Gene Silencing and Cause Genome-Wide Reductions in Cytosine Methylation. J. Virol. 2009, 83, 5005–5013. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Wang, H.; Sunter, G.; Bisaro, D.M. Geminivirus AL2 and L2 Proteins Interact with and Inactivate SNF1 Kinase. Plant Cell 2003, 15, 1034–1048. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Negrete, E.; Lozano-Durán, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef] [Green Version]
- Castillo-González, C.; Liu, X.; Huang, C.; Zhao, C.; Ma, Z.; Hu, T.; Sun, F.; Zhou, Y.; Zhou, X.; Wang, X.-J.; et al. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 2015, 4, 1–31. [Google Scholar] [CrossRef]
- Brough, C.L.; Gardiner, W.E.; Inamdar, N.M.; Zhang, X.Y.; Ehrlich, M.; Bisaro, D.M. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 1992, 18, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Coursey, T.; Regedanz, E.; Bisaro, D.M. Arabidopsis RNA Polymerase V Mediates Enhanced Compaction and Silencing of Geminivirus and Transposon Chromatin during Host Recovery from Infection. J. Virol. 2018, 92, e01320-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral Genome Methylation as an Epigenetic Defense against Geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis Double-Stranded RNA Binding Protein DRB3 Participates in Methylation-Mediated Defense against Geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackel, J.N.; Storer, J.M.; Coursey, T.; Bisaro, D.M. Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin. J. Virol. 2016, 90, 7529–7540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagen, C.; Rojas, M.R.; Kon, T.; Gilbertson, R.L. Recovery from Cucurbit leaf crumple virus (Family Geminiviridae, Genus Begomovirus) infection is an adaptive antiviral response associated with changes in viral small RNAs. Phytopathology 2008, 98, 1029–1037. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.T.; Voinnet, O.; Baulcombe, D.C. Initiation and Maintenance of Virus-Induced Gene Silencing. Plant Cell 1998, 10, 937–946. [Google Scholar] [CrossRef]
- Jones, L.; Hamilton, A.J.; Voinnet, O.; Thomas, C.L.; Maule, A.J.; Baulcombe, D.C. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 1999, 11, 2291–2301. [Google Scholar]
- Bonilla-Ramírez, G.M.; Guevara-González, R.G.; Garzón-Tiznado, J.A.; Ascencio-Ibáñez, J.T.; Torres-Pacheco, I.; Rivera-Bustamante, R.F. Analysis of the infectivity of monomeric clones of pepper huasteco virus. J. Gen. Virol. 1997, 78, 947–951. [Google Scholar] [CrossRef]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 1992, 2, 549–557. [Google Scholar]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Peart, J.; Wu, A.-J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003, 22, 5690–5699. [Google Scholar] [CrossRef] [PubMed]
- Cañizares, M.C.; Rosas-Díaz, T.; Rodríguez-Negrete, E.; Hogenhout, S.A.; Bedford, I.D.; Bejarano, E.R.; Navas-Castillo, J.; Moriones, E. Arabidopsis thaliana, an experimental host for tomato yellow leaf curl disease-associated begomoviruses by agroinoculation and whitefly transmission. Plant Pathol. 2015, 64, 265–271. [Google Scholar] [CrossRef]
- Lee, H.A.; Kim, S.Y.; Oh, S.K.; Yeom, S.I.; Kim, S.B.; Kim, M.S.; Kamoun, S.; Choi, D. Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 2014, 203, 926–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb. Protoc. 2017, 2017, pdb.prot093450. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, F.; Huang, C.; Yang, X.; Qian, Y.; Xie, Y.; Zhou, X. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J. Gen. Virol. 2014, 95, 225–230. [Google Scholar] [CrossRef]
- Li, F.; Xu, X.; Huang, C.; Gu, Z.; Cao, L.; Hu, T.; Ding, M.; Li, Z.; Zhou, X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef] [Green Version]
- Gruntman, E.; Qi, Y.; Slotkin, R.K.; Roeder, T.; Martienssen, R.A.; Sachidanandam, R. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 2008, 9, 371. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.; Alvarez-Venegas, R.; Avramova, Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008, 3, 1018–1025. [Google Scholar] [CrossRef]
- Lakatos, L.; Szittya, G.; Silhavy, D.; Burgyán, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004, 23, 876–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roossinck, M.J. Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 2005, 3, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Argüello-Astorga, G.R.; Guevara-González, R.G.; Herrera-Estrella, L.R.; Rivera-Bustamante, R.F. Geminivirus Replication Origins Have a Group-Specific Organization of Iterative Elements: A Model for Replication. Virology 1994, 203, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Shin, S.H.; Jeon, E.M.; Park, J.M.; Hyun, J.Y.; Harn, C.H. Pepper, Chili (Capsicum annuum). In Methods in Molecular Biology (Clifton, N.J.); Springer: New York, NY, USA, 2015; Volume 1223, pp. 311–320. ISBN 9781493916955. [Google Scholar]
- Jackel, J.N.; Buchmann, R.C.; Singhal, U.; Bisaro, D.M. Analysis of Geminivirus AL2 and L2 Proteins Reveals a Novel AL2 Silencing Suppressor Activity. J. Virol. 2015, 89, 3176–3187. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Baliji, S.; Buchmann, R.C.; Wang, H.; Lindbo, J.A.; Sunter, G.; Bisaro, D.M. Functional Modulation of the Geminivirus AL2 Transcription Factor and Silencing Suppressor by Self-Interaction. J. Virol. 2007, 81, 11972–11981. [Google Scholar] [CrossRef] [Green Version]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [Green Version]
- Zhan, B.; Zhao, W.; Li, S.; Yang, X.; Zhou, X. Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing. Viruses 2018, 10, 488. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Ren, Y.; Sun, S.; Wang, D.; Zhang, F.; Li, D.; Li, S.; Zhou, X. Identification of the potential virulence factors and RNA silencing suppressors of mulberry mosaic dwarf-associated geminivirus. Viruses 2018, 10, 472. [Google Scholar] [CrossRef] [Green Version]
- Luna, A.P.; Rodríguez-Negrete, E.A.; Morilla, G.; Wang, L.; Lozano-Durán, R.; Castillo, A.G.; Bejarano, E.R. V2 from a curtovirus is a suppressor of post-transcriptional gene silencing. J. Gen. Virol. 2017, 98, 2607–2614. [Google Scholar] [CrossRef]
- Garzon-Tiznado, J.A.; Torres-Pacheco, I.; Ascencio-Ibáñez, J.T.; Herrera-Estrella, L.; Rivera-Bustamante, R.F. Inoculation of Peppers with Infectious Clones of a New Geminivirus by a Biolistic Procedure. Phytopathology 1993, 83, 514. [Google Scholar] [CrossRef]
- Sunter, G.; Stenger, D.C.; Bisaro, D.M. Heterologous Complementation by Geminivirus AL2 and AL3 Genes. Virology 1994, 203, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Settlage, S.B.; See, R.G.; Hanley-Bowdoin, L. Geminivirus C3 Protein: Replication Enhancement and Protein Interactions. J. Virol. 2005, 79, 9885–9895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, A.G.; Collinet, D.; Deret, S.; Kashoggi, A.; Bejarano, E.R. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 2003, 312, 381–394. [Google Scholar] [CrossRef]
- Singh, D.K.; Islam, M.N.; Choudhury, N.R.; Karjee, S.; Mukherjee, S.K. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 2007, 35, 755–770. [Google Scholar] [CrossRef]
- Pasumarthy, K.K.; Mukherjee, S.K.; Choudhury, N.R. The presence of tomato leaf curl Kerala virus AC3 protein enhances viral DNA replication and modulates virus induced gene-silencing mechanism in tomato plants. Virol. J. 2011, 8, 178. [Google Scholar] [CrossRef] [Green Version]
- Elmayan, T.; Proux, F.; Vaucheret, H. Arabidopsis RPA2: A genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 2005, 15, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Xia, R.; Wang, J.; Liu, C.; Wang, Y.; Wang, Y.; Zhai, J.; Liu, J.; Hong, X.; Cao, X.; Zhu, J.K.; et al. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 2006, 18, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Czosnek, H.; Eybishtz, A.; Sade, D.; Gorovits, R.; Sobol, I.; Bejarano, E.; Rosas-Díaz, T.; Lozano-Durán, R. Discovering Host Genes Involved in the Infection by the Tomato Yellow Leaf Curl Virus Complex and in the Establishment of Resistance to the Virus Using Tobacco Rattle Virus-based Post Transcriptional Gene Silencing. Viruses 2013, 5, 998–1022. [Google Scholar] [CrossRef] [Green Version]
- Krenz, B.; Deuschle, K.; Deigner, T.; Unseld, S.; Kepp, G.; Wege, C.; Kleinow, T.; Jeske, H. Early Function of the Abutilon Mosaic Virus AC2 Gene as a Replication Brake. J. Virol. 2015, 89, 3683–3699. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ding, X.; Xiao, J.; Jiménez-Gόngora, T.; Liu, R.; Lozano-Durán, R. Inference of a geminivirus−host protein−protein interaction network through affinity purification and mass spectrometry analysis. Viruses 2017, 9, 275. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Gandarilla, M.G.; Rodríguez-Negrete, E.A.; Rivera-Bustamante, R.F. Superinfection by PHYVV Alters the Recovery Process in PepGMV-Infected Pepper Plants. Viruses 2020, 12, 286. https://doi.org/10.3390/v12030286
Rodríguez-Gandarilla MG, Rodríguez-Negrete EA, Rivera-Bustamante RF. Superinfection by PHYVV Alters the Recovery Process in PepGMV-Infected Pepper Plants. Viruses. 2020; 12(3):286. https://doi.org/10.3390/v12030286
Chicago/Turabian StyleRodríguez-Gandarilla, Myriam G., Edgar A. Rodríguez-Negrete, and Rafael F. Rivera-Bustamante. 2020. "Superinfection by PHYVV Alters the Recovery Process in PepGMV-Infected Pepper Plants" Viruses 12, no. 3: 286. https://doi.org/10.3390/v12030286
APA StyleRodríguez-Gandarilla, M. G., Rodríguez-Negrete, E. A., & Rivera-Bustamante, R. F. (2020). Superinfection by PHYVV Alters the Recovery Process in PepGMV-Infected Pepper Plants. Viruses, 12(3), 286. https://doi.org/10.3390/v12030286