The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take
Abstract
1. Introduction
2. Rotavirus Classification and Genotypes
3. In Vitro Binding Studies Show Rotavirus Binding to HBGAs in a P Genotype–Dependent Manner
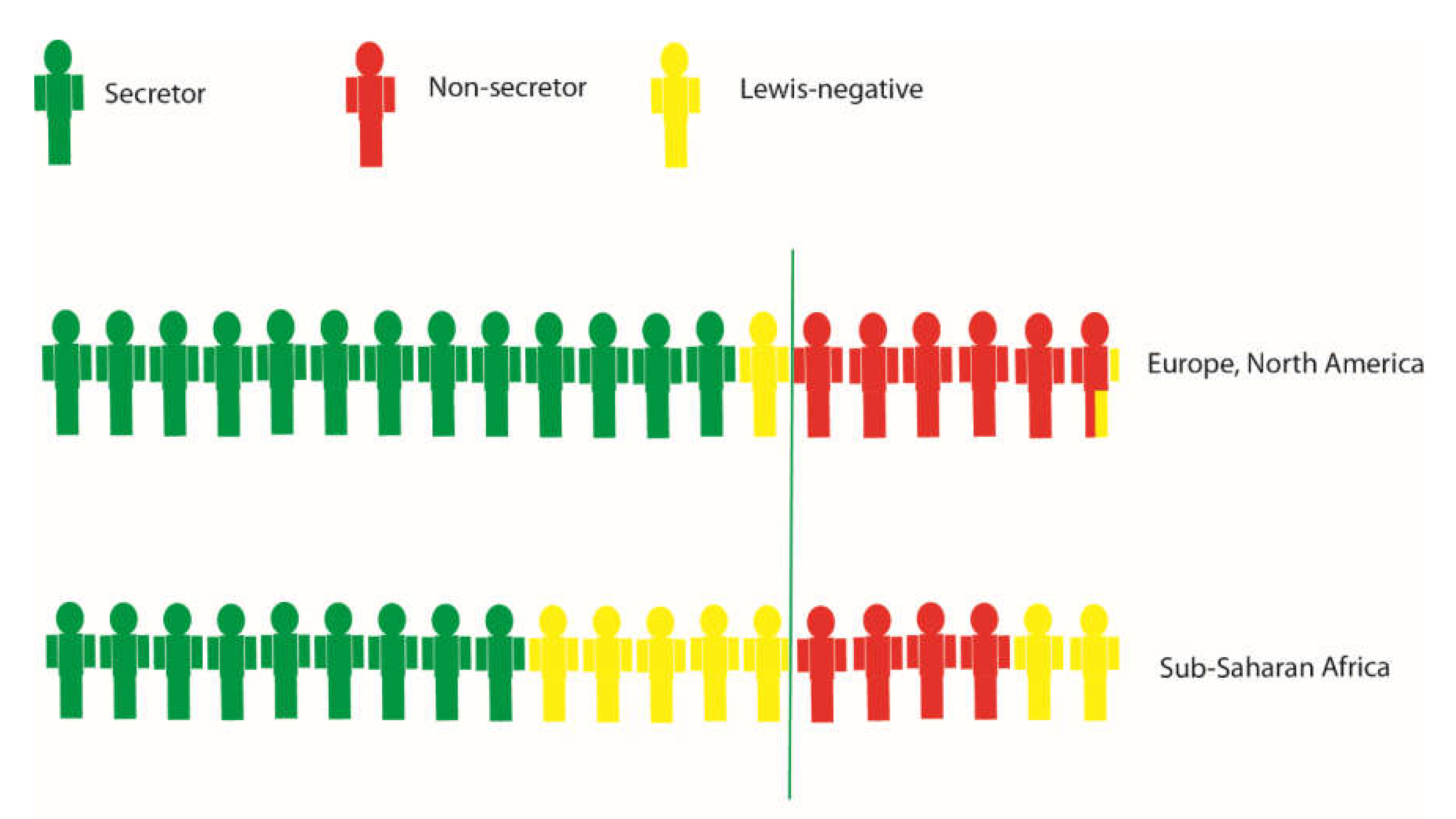
4. The HBGA Biosynthesis Pathway
5. Rotavirus Susceptibility In Vivo Is Strongly Associated with HBGAs in a P Genotype–Dependent Manner
6. Secretor-Positive Adults Have Significantly Higher Immunoglobulin G (IgG), IgA, and Neutralization Antibody Titers to Rotavirus Compared to Non-Secretors
7. Population Genetics and Epidemiology of Rotaviruses
8. Zoonosis and Host Genetics
9. Neonatal Infections and Host Genetics
10. Rotavirus Vaccine Take Is Associated with HBGAs
11. Will Reduced Vaccine Take Translate to More Clinical Vaccine Failures?
12. Human Intestinal Enteroids: A Novel Model for Studying Genetic Susceptibility to Rotavirus Infections
13. Conclusions and Outstanding Questions
Author Contributions
Funding
Conflicts of Interest
References
- Shepherd, F.K.; Herrera-Ibata, D.M.; Porter, E.; Homwong, N.; Hesse, R.; Bai, J.; Marthaler, D.G. Whole genome classification and phylogenetic analyses of rotavirus b strains from the United States. Pathogens 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Doro, R.; Laszlo, B.; Martella, V.; Leshem, E.; Gentsch, J.; Parashar, U.; Banyai, K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: Is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 2014, 28, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Banyai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (rcwg). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, P.; Tan, M.; Liu, Y.; Biesiada, J.; Meller, J.; Castello, A.A.; Jiang, B.; Jiang, X. Rotavirus vp8*: Phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012, 86, 9899–9910. [Google Scholar] [CrossRef] [PubMed]
- Isa, P.; Arias, C.F.; Lopez, S. Role of sialic acids in rotavirus infection. Glycoconj. J. 2006, 23, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Czako, R.; Cortes-Penfield, N.W.; Smith, D.F.; Le Pendu, J.; Estes, M.K.; Prasad, B.V. Cell attachment protein vp8* of a human rotavirus specifically interacts with a-type histo-blood group antigen. Nature 2012, 485, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein vp8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843. [Google Scholar] [CrossRef]
- Ramani, S.; Cortes-Penfield, N.W.; Hu, L.; Crawford, S.E.; Czako, R.; Smith, D.F.; Kang, G.; Ramig, R.F.; Le Pendu, J.; Prasad, B.V.; et al. The vp8* domain of neonatal rotavirus strain g10p[11] binds to type ii precursor glycans. J. Virol. 2013, 87, 7255–7264. [Google Scholar] [CrossRef]
- Barbe, L.; Le Moullac-Vaidye, B.; Echasserieau, K.; Bernardeau, K.; Carton, T.; Bovin, N.; Nordgren, J.; Svensson, L.; Ruvoen-Clouet, N.; Le Pendu, J. Histo-blood group antigen-binding specificities of human rotaviruses are associated with gastroenteritis but not with in vitro infection. Sci. Rep. 2018, 8, 12961. [Google Scholar] [CrossRef]
- Sun, X.; Guo, N.; Li, D.; Jin, M.; Zhou, Y.; Xie, G.; Pang, L.; Zhang, Q.; Cao, Y.; Duan, Z.J. Binding specificity of p[8] vp8* proteins of rotavirus vaccine strains with histo-blood group antigens. Virology 2016, 495, 129–135. [Google Scholar] [CrossRef]
- Gozalbo-Rovira, R.; Ciges-Tomas, J.R.; Vila-Vicent, S.; Buesa, J.; Santiso-Bellon, C.; Monedero, V.; Yebra, M.J.; Marina, A.; Rodriguez-Diaz, J. Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens. PLoS Pathog. 2019, 15, e1007865. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, J.; Sharma, S.; Bucardo, F.; Nasir, W.; Gunaydin, G.; Ouermi, D.; Nitiema, L.W.; Becker-Dreps, S.; Simpore, J.; Hammarstrom, L.; et al. Both lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin. Infect. Dis. 2014, 59, 1567–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Long, Y.; Tan, M.; Zhang, T.; Huang, Q.; Jiang, X.; Tan, W.F.; Li, J.D.; Hu, G.F.; Tang, S.; et al. P[8] and p[4] rotavirus infection associated with secretor phenotypes among children in south china. Sci. Rep. 2016, 6, 34591. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guo, N.; Li, J.; Yan, X.; He, Z.; Li, D.; Jin, M.; Xie, G.; Pang, L.; Zhang, Q.; et al. Rotavirus infection and histo-blood group antigens in the children hospitalized with diarrhoea in china. Clin. Microbiol. Infect. 2016, 22, 740–e741. [Google Scholar] [CrossRef] [PubMed]
- Pollock, L.; Bennett, A.; Jere, K.C.; Dube, Q.; Mandolo, J.; Bar-Zeev, N.; Heyderman, R.S.; Cunliffe, N.A.; Iturriza-Gomara, M. Non-secretor histo-blood group antigen phenotype is associated with reduced risk of clinical rotavirus vaccine failure in malawian infants. Clin. Infect. Dis. 2018, 69, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Dickson, D.M.; de Camp, A.C.; Ross Colgate, E.; Diehl, S.A.; Uddin, M.I.; Sharmin, S.; Islam, S.; Bhuiyan, T.R.; Alam, M.; et al. Histo-blood group antigen phenotype determines susceptibility to genotype-specific rotavirus infections and impacts measures of rotavirus vaccine efficacy. J. Infect. Dis. 2018, 217, 1399–1407. [Google Scholar] [CrossRef]
- Van Trang, N.; Vu, H.T.; Le, N.T.; Huang, P.; Jiang, X.; Anh, D.D. Association between norovirus and rotavirus infection and histo-blood group antigen types in vietnamese children. J. Clin. Microbiol. 2014, 52, 1366–1374. [Google Scholar] [CrossRef]
- Payne, D.C.; Currier, R.L.; Staat, M.A.; Sahni, L.C.; Selvarangan, R.; Halasa, N.B.; Englund, J.A.; Weinberg, G.A.; Boom, J.A.; Szilagyi, P.G.; et al. Epidemiologic association between fut2 secretor status and severe rotavirus gastroenteritis in children in the united states. JAMA Pediatr. 2015, 169, 1040–1045. [Google Scholar] [CrossRef]
- Perez-Ortin, R.; Vila-Vicent, S.; Carmona-Vicente, N.; Santiso-Bellon, C.; Rodriguez-Diaz, J.; Buesa, J. Histo-blood group antigens in children with symptomatic rotavirus infection. Viruses 2019, 11. [Google Scholar] [CrossRef]
- Imbert-Marcille, B.M.; Barbe, L.; Dupe, M.; Le Moullac-Vaidye, B.; Besse, B.; Peltier, C.; Ruvoen-Clouet, N.; Le Pendu, J. A fut2 gene common polymorphism determines resistance to rotavirus a of the p[8] genotype. J. Infect. Dis. 2014, 209, 1227–1230. [Google Scholar] [CrossRef]
- Ayouni, S.; Sdiri-Loulizi, K.; de Rougemont, A.; Estienney, M.; Ambert-Balay, K.; Aho, S.; Hamami, S.; Aouni, M.; Neji-Guediche, M.; Pothier, P.; et al. Rotavirus p[8] infections in persons with secretor and nonsecretor phenotypes, tunisia. Emerg. Infect. Dis. 2015, 21, 2055–2058. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, J.; Svensson, L. Genetic susceptibility to human norovirus infection: An update. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Gunaydin, G.; Nordgren, J.; Sharma, S.; Hammarstrom, L. Association of elevated rotavirus-specific antibody titers with hbga secretor status in Swedish individuals: The fut2 gene as a putative susceptibility determinant for infection. Virus Res. 2016, 211, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Diaz, J.; Garcia-Mantrana, I.; Vila-Vicent, S.; Gozalbo-Rovira, R.; Buesa, J.; Monedero, V.; Collado, M.C. Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans. Sci. Rep. 2017, 7, 45559. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, J.; Sharma, S.; Kambhampati, A.; Lopman, B.; Svensson, L. Innate resistance and susceptibility to norovirus infection. PLoS Pathog. 2016, 12, e1005385. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Kindberg, E.; Paniagua, M.; Grahn, A.; Larson, G.; Vildevall, M.; Svensson, L. Genetic susceptibility to symptomatic norovirus infection in nicaragua. J. Med. Virol. 2009, 81, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Reyes, Y.; Ronnelid, Y.; Gonzalez, F.; Sharma, S.; Svensson, L.; Nordgren, J. Histo-blood group antigens and rotavirus vaccine shedding in nicaraguan infants. Sci. Rep. 2019, 9, 10764. [Google Scholar] [CrossRef]
- Piedade, J.; Nordgren, J.; Esteves, F.; Esteves, A.; Teodosio, R.; Svensson, L.; Istrate, C. Molecular epidemiology and host genetics of norovirus and rotavirus infections in portuguese elderly living in aged care homes. J. Med. Virol. 2019, 91, 1014–1021. [Google Scholar] [CrossRef]
- Todd, S.; Page, N.A.; Duncan Steele, A.; Peenze, I.; Cunliffe, N.A. Rotavirus strain types circulating in africa: Review of studies published during 1997–2006. J. Infect. Dis. 2010, 202 (Suppl. 1), S34–S42. [Google Scholar] [CrossRef]
- Yen, C.; Steiner, C.A.; Barrett, M.; Curns, A.T.; Hunter, K.; Wilson, E.; Parashar, U.D. Racial disparities in diarrhea-associated hospitalizations among children in five us states, before and after introduction of rotavirus vaccine. Vaccine 2010, 28, 7423–7426. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Tan, M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect. 2017, 6, e22. [Google Scholar] [CrossRef]
- Wenske, O.; Ruckner, A.; Piehler, D.; Schwarz, B.A.; Vahlenkamp, T.W. Epidemiological analysis of porcine rotavirus a genotypes in germany. Vet. Microbiol. 2018, 214, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Gonzalez, F.; Reyes, Y.; Blandon, P.; Saif, L.; Nordgren, J. Seroprevalence in household raised pigs indicate high exposure to gii noroviruses in rural nicaragua. Zoonoses. Public Health 2016, 63, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Jiang, X.; Zhong, W.; Jensen, H.M.; Brandl, M.; Bates, A.H.; Engelbrektson, A.L.; Mandrell, R. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res. Vet. Sci. 2007, 83, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; McGregor, R.; Meulia, T.; Wang, Q.; Saif, L.J. Binding patterns of human norovirus-like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to a/h histo-blood group antigen expression. J. Virol. 2007, 81, 3535–3544. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ramelot, T.A.; Huang, P.; Liu, Y.; Li, Z.; Feizi, T.; Zhong, W.; Wu, F.T.; Tan, M.; Kennedy, M.A.; et al. Glycan specificity of p[19] rotavirus and comparison with those of related p genotypes. J. Virol. 2016, 90, 9983–9996. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, S.; Woodruff, A.L.; Xia, M.; Tan, M.; Kennedy, M.A.; Jiang, X. Structural basis of glycan specificity of p[19] vp8*: Implications for rotavirus zoonosis and evolution. PLoS Pathog. 2017, 13, e1006707. [Google Scholar] [CrossRef]
- Sun, X.; Li, D.; Qi, J.; Chai, W.; Wang, L.; Wang, L.; Peng, R.; Wang, H.; Zhang, Q.; Pang, L.; et al. Glycan binding specificity and mechanism of human and porcine p[6]/p[19] rotavirus vp8*s. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Potgieter, C.A.; Ciarlet, M.; Parreno, V.; Martella, V.; Banyai, K.; Garaicoechea, L.; Palombo, E.A.; Novo, L.; Zeller, M.; et al. Are human p[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order artiodactyla? J. Virol. 2009, 83, 2917–2929. [Google Scholar] [CrossRef]
- Midgley, S.E.; Hjulsager, C.K.; Larsen, L.E.; Falkenhorst, G.; Bottiger, B. Suspected zoonotic transmission of rotavirus group a in danish adults. Epidemiol. Infect. 2012, 140, 1013–1017. [Google Scholar] [CrossRef]
- Bohm, R.; Fleming, F.E.; Maggioni, A.; Dang, V.T.; Holloway, G.; Coulson, B.S.; von Itzstein, M.; Haselhorst, T. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 2015, 6, 5907. [Google Scholar] [CrossRef] [PubMed]
- Haffejee, I.E. Neonatal rotavirus infections. Rev. Infect. Dis 1991, 13, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Iturriza Gomara, M.; Kang, G.; Mammen, A.; Jana, A.K.; Abraham, M.; Desselberger, U.; Brown, D.; Gray, J. Characterization of g10p[11] rotaviruses causing acute gastroenteritis in neonates and infants in vellore, india. J. Clin. Microbiol. 2004, 42, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Choi, S.; Shin, S.H.; Lee, E.J.; Hyun, J.; Kim, J.S.; Kim, H.S. Emergence of g8p[6] rotavirus strains in korean neonates. Gut Pathog. 2018, 10, 27. [Google Scholar] [CrossRef]
- Mascarenhas, J.D.; Linhares, A.C.; Gabbay, Y.B.; Lima, C.S.; Guerra Sde, F.; Soares, L.S.; Oliveira, D.S.; Lima, J.C.; Macedo, O.; Leite, J.P. Molecular characterization of vp4 and nsp4 genes from rotavirus strains infecting neonates and young children in belem, brazil. Virus Res. 2007, 126, 149–158. [Google Scholar] [CrossRef]
- Ray, P.; Sharma, S.; Agarwal, R.K.; Longmei, K.; Gentsch, J.R.; Paul, V.K.; Glass, R.I.; Bhan, M.K. First detection of g12 rotaviruses in newborns with neonatal rotavirus infection at all india institute of medical sciences, new delhi, india. J. Clin. Microbiol. 2007, 45, 3824–3827. [Google Scholar] [CrossRef][Green Version]
- Hu, L.; Sankaran, B.; Laucirica, D.R.; Patil, K.; Salmen, W.; Ferreon, A.C.M.; Tsoi, P.S.; Lasanajak, Y.; Smith, D.F.; Ramani, S.; et al. Glycan recognition in globally dominant human rotaviruses. Nat. Commun. 2018, 9, 2631. [Google Scholar] [CrossRef]
- Ramani, S.; Hu, L.; Venkataram Prasad, B.V.; Estes, M.K. Diversity in rotavirus-host glycan interactions: A “sweet” spectrum. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 263–273. [Google Scholar] [CrossRef]
- Karlsson, K.A.; Larson, G. Molecular characterization of cell surface antigens of fetal tissue. Detailed analysis of glycosphingolipids of meconium of a human o le(a--b+) secretor. J. Biol. Chem. 1981, 256, 3512–3524. [Google Scholar]
- Clark, A.; van Zandvoort, K.; Flasche, S.; Sanderson, C.; Bines, J.; Tate, J.; Parashar, U.; Jit, M. Efficacy of live oral rotavirus vaccines by duration of follow-up: A meta-regression of randomised controlled trials. Lancet Infect. Dis. 2019, 19, 717–727. [Google Scholar] [CrossRef]
- Sharma, S.; Nordgren, J. Rotavirus vaccines in developing countries: Issues and future considerations. Future Virol. 2015, 10, 663–666. [Google Scholar] [CrossRef]
- Steele, A.D.; Victor, J.C.; Carey, M.E.; Tate, J.E.; Atherly, D.E.; Pecenka, C.; Diaz, Z.; Parashar, U.D.; Kirkwood, C.D. Experiences with rotavirus vaccines: Can we improve rotavirus vaccine impact in developing countries? Hum. Vaccin. Immunother. 2019, 15, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Nordgren, J.; Reyes, Y.; Gonzalez, F.; Sharma, S.; Svensson, L. The lewis a phenotype is a restriction factor for rotateq and rotarix vaccine-take in nicaraguan children. Sci. Rep. 2018, 8, 1502. [Google Scholar] [CrossRef] [PubMed]
- Kazi, A.M.; Cortese, M.M.; Yu, Y.; Lopman, B.; Morrow, A.L.; Fleming, J.A.; McNeal, M.M.; Steele, A.D.; Parashar, U.D.; Zaidi, A.K.M.; et al. Secretor and salivary abo blood group antigen status predict rotavirus vaccine take in infants. J. Infect. Dis. 2017, 215, 786–789. [Google Scholar] [CrossRef]
- Armah, G.E.; Cortese, M.M.; Dennis, F.E.; Yu, Y.; Morrow, A.L.; McNeal, M.M.; Lewis, K.D.C.; Awuni, D.A.; Armachie, J.; Parashar, U.D. Rotavirus vaccine take in infants is associated with secretor status. J. Infect. Dis. 2019, 219, 746–749. [Google Scholar] [CrossRef]
- Boniface, K.; Byars, S.G.; Cowley, D.; Kirkwood, C.D.; Bines, J.E. Human neonatal rotavirus vaccine (rv3-bb) produces vaccine take irrespective of histo-blood group antigen status. J. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Bjork, S.; Breimer, M.E.; Hansson, G.C.; Karlsson, K.A.; Leffler, H. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the abo, le and se phenotype of the donor. J. Biol. Chem. 1987, 262, 6758–6765. [Google Scholar]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef]
- Costantini, V.; Morantz, E.K.; Browne, H.; Ettayebi, K.; Zeng, X.L.; Atmar, R.L.; Estes, M.K.; Vinje, J. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 2018, 24, 1453–1464. [Google Scholar] [CrossRef]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human intestinal enteroids: A new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef]


| P-Genotype | Country | Lewis | ABO | Secretor | Reference |
|---|---|---|---|---|---|
| P[4] | |||||
| Nicaragua | Only Lewis b infected | Not investigated | Only secretors infected | [12] | |
| China | Lewis a less infected | No association | Secretors more infected | [13] | |
| China | No association | No association | Only secretors/partial secretors infected | [14] | |
| Malawi | Lewis negative less susceptible | No association | Secretors more infected | [15] | |
| Bangladesh | Only Lewis b infected | Not investigated | Only secretors infected | [16] | |
| Vietnam | Lewis b present in all infected | No association | Only secretors/partial secretors infected | [17] | |
| USA | Not investigated | Not investigated | Only secretors infected | [18] | |
| P[6] | |||||
| Burkina Faso | Lewis negative more susceptible | No association | No association | [12] | |
| Malawi | Lewis negative more susceptible | No association | No association | [15] | |
| Bangladesh | Lewis negative more susceptible | Not investigated | No association | [16] | |
| Vietnam | Lewis negative more susceptible a | No association | Not investigated | [17] | |
| P[8] | |||||
| Burkina Faso | Only Lewis b infected | No association | Only secretors infected | [12] | |
| Nicaragua | Only Lewis b infected | Not investigated | Only secretors infected | [12] | |
| China | Lewis a less infected | No association | Secretors more infected | [13] | |
| China | Lewis a less infected | No association | Secretors more infected | [14] | |
| Spain | Lewis b more infected | Blood group A and AB more infected compared to O | Secretors more infected | [19] | |
| Malawi | Lewis negative less susceptible | No association | Secretors more infected | [15] | |
| Bangladesh | Lewis negative less susceptible | Not investigated | No association | [16] | |
| Vietnam | No Lewis a infected | No association | Only secretors/partial secretors infected | [17] | |
| France | Not investigated | Not investigated | Only secretors infected | [20] | |
| USA | Not investigated | Not investigated | Apart from 1, only secretors infected | [18] | |
| Tunisia | Only Lewis positives infected | No association | No association | [21] |
| Country | Type of Response | ABO | Lewis | Secretor Status | Reference |
|---|---|---|---|---|---|
| Sweden | Serum IgG | Not investigated | No association a | Secretors higher titers to rotavirus | [23] |
| Sweden | Neutralization antibody titers | Not investigated | No association | Secretors higher neutralization antibody titers to P[8] but not P[6] | [23] |
| France | Neutralization antibody titers | Not investigated | Not investigated | Secretors higher neutralization titers to P[8] | [9] |
| Spain | Salivary IgA | Not investigated | Not investigated | Secretors higher titers to rotavirus | [24] |
| China | Serum IgG | No association | No association b | Secretors higher titers to VP8* of genotype P[4] and P[8] | [13] |
| Vaccine | Country | ABO | Lewis | Secretor Status | Method | Reference | |
|---|---|---|---|---|---|---|---|
| Time Point | Measurement | ||||||
| Rotarix | Nicaragua | B less seroconversion a | Lewis A no seroconversion | Non-secretors less seroconversion b | After 1 dose | [53] | |
| Pakistan | Non-O less seroconversion compared to O | No association | Non-secretors less seroconversion | After 3 doses | [54] | ||
| Ghana | O less seroconversion compared to B | No association | Non-secretors less seroconversion | After 2-3 doses | Seroconversion | [55] | |
| Malawi | No association | No association | Non-secretors less seroconversion b | After 2 doses | [15] | ||
| RotaTeq | Nicaragua | A had most seroconversion a | Lewis A no seroconversion, low power | No association | After 1 dose | [53] | |
| RV3-BB | New Zealand | Not investigated | No association, low power | No association | Cumulative | [56] | |
| Rotarix | Nicaragua | B no shedding, low power | Lewis A no shedding, low power | Non-secretors no shedding b | After 1 dose | [27] | |
| Malawi | No association | No association | Non-secretors less shedding | After 1 dose | Vaccine shedding | [15] | |
| Malawi | No association | No association | No association | After 2 doses | [15] | ||
| RotaTeq | Nicaragua | No association | Lewis A no shedding, low power | No association | After 1 dose | [27] | |
| RV3-BB | New Zealand | Not investigated | No association, low power | No association | Cumulative | [56] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Hagbom, M.; Svensson, L.; Nordgren, J. The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take. Viruses 2020, 12, 324. https://doi.org/10.3390/v12030324
Sharma S, Hagbom M, Svensson L, Nordgren J. The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take. Viruses. 2020; 12(3):324. https://doi.org/10.3390/v12030324
Chicago/Turabian StyleSharma, Sumit, Marie Hagbom, Lennart Svensson, and Johan Nordgren. 2020. "The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take" Viruses 12, no. 3: 324. https://doi.org/10.3390/v12030324
APA StyleSharma, S., Hagbom, M., Svensson, L., & Nordgren, J. (2020). The Impact of Human Genetic Polymorphisms on Rotavirus Susceptibility, Epidemiology, and Vaccine Take. Viruses, 12(3), 324. https://doi.org/10.3390/v12030324





