High Transmission Potential of West Nile Virus Lineage 1 for Cx. pipiens s.l. of Iran
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethic Statements
2.2. Culex Pipiens Collection and Rearing
2.3. Vector Competence Assay
2.4. Statistical Analysis
3. Results
3.1. Artificial Feeding
3.2. Viral Infection Over Time
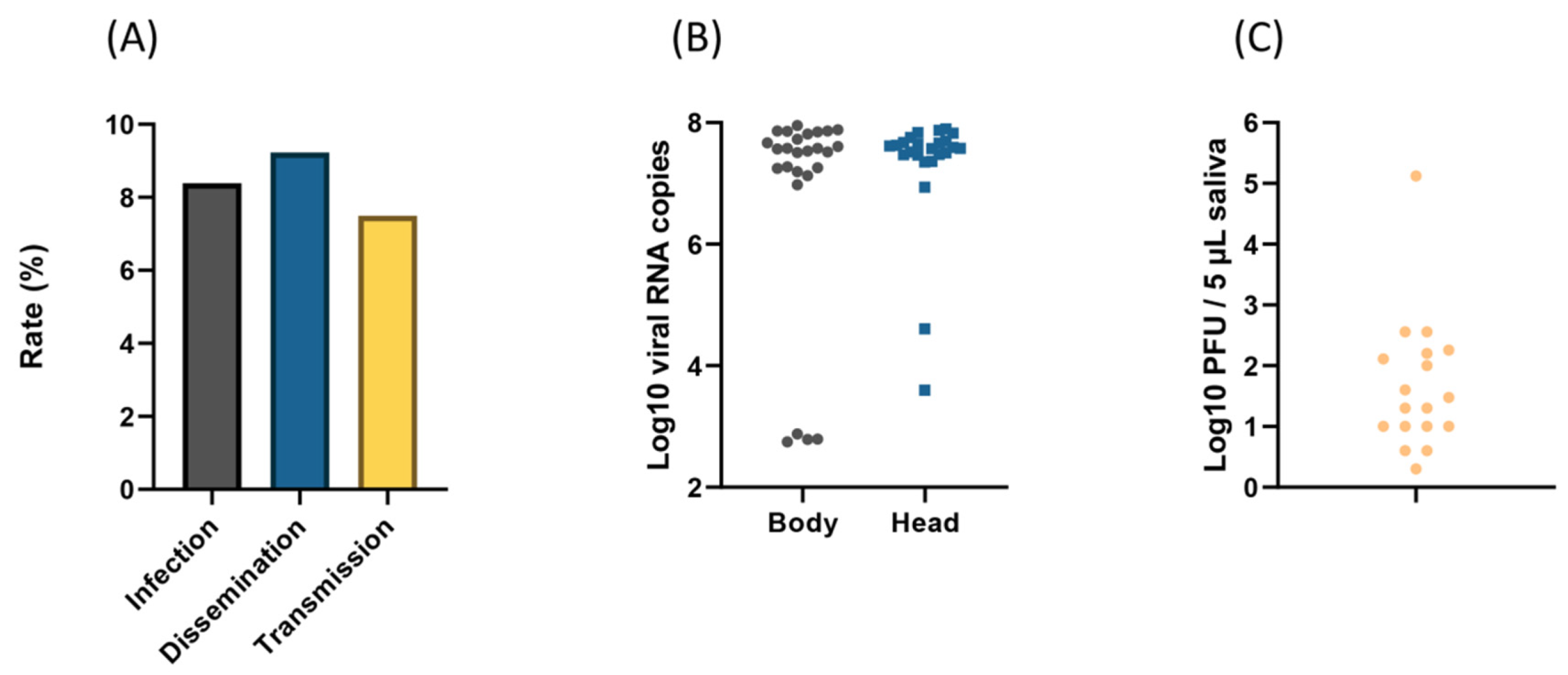
3.3. Dissemination and Transmission at 21 Days Post-Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caraballo, H.; King, K. Emergency department management of mosquito-borne illness: Malaria, dengue, and West Nile virus. Emerg. Med. Pract. 2014, 16, 1–23. [Google Scholar] [PubMed]
- Kramer, L.D.; Styer, L.M.; Ebel, G.D. A global perspective on the epidemiology of West Nile virus. Annu. Rev. Entomol. 2008, 53, 61–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smithburn, K.; Hughes, T.; Burke, A.; Paul, J. A neurotropic virus isolated from the blood of a native of Uganda1. Am. J. Trop. Med. Hyg. 1940, 1, 471–492. [Google Scholar] [CrossRef]
- Andreadis, T.G. The contribution of Culex pipiens complex mosquitoes to transmission and persistence of West Nile virus in North America. J. Am. Mosq. Control. Assoc. 2012, 28, 137–152. [Google Scholar] [CrossRef]
- Conley, A.K.; Fuller, D.O.; Haddad, N.; Hassan, A.N.; Gad, A.M.; Beier, J.C. Modeling the distribution of the West Nile and Rift Valley Fever vector Culex pipiens in arid and semi-arid regions of the Middle East and North Africa. Parasit. Vectors 2014, 7, 289. [Google Scholar] [CrossRef] [Green Version]
- Petersen, L.R.; Roehrig, J.T.; Sejvar, J.J. West Nile virus in the Americas. In New and Evolving Infections of the 21st Century; Springer: Berlin/Heidelberg, Germany, 2007; pp. 3–56. [Google Scholar]
- Rizzoli, A.; Jimenez-Clavero, M.; Barzon, L.; Cordioli, P.; Figuerola, J.; Koraka, P.; Martina, B.; Moreno, A.; Nowotny, N.; Pardigon, N. The challenge of West Nile virus in Europe: Knowledge gaps and research priorities. Euro. Surveill. 2015, 20, 21135. [Google Scholar] [CrossRef] [Green Version]
- Heydari, M.; Metanat, M.; Rouzbeh-Far, M.-A.; Tabatabaei, S.M.; Rakhshani, M.; Sepehri-Rad, N.; Keshtkar-Jahromi, M. Dengue Fever as an Emerging Infection in Southeast Iran. Am. J. Trop. Med. Hyg. 2018, 98, 1469–1471. [Google Scholar] [CrossRef] [Green Version]
- Ziyaeyan, M.; Behzadi, M.A.; Leyva-Grado, V.H.; Azizi, K.; Pouladfar, G.; Dorzaban, H.; Ziyaeyan, A.; Salek, S.; Hashemi, A.J.; Jamalidoust, M. Widespread circulation of West Nile virus, but not Zika virus in southern Iran. PLoS Negl. Trop. Dis. 2018, 12, e0007022. [Google Scholar] [CrossRef]
- Naficy, K.; Saidi, S. Serological survey on viral antibodies in Iran. Trop. Geogr. Med. 1970, 22, 183–188. [Google Scholar]
- Saidi, S.; Tesh, R.; Javadian, E.; Nadim, A. The prevalence of human infection with West Nile virus in Iran. Iran. J. Public Health 1976, 5, 8–13. [Google Scholar]
- Zou, S.; Foster, G.A.; Dodd, R.Y.; Petersen, L.R.; Stramer, S.L. West Nile fever characteristics among viremic persons identified through blood donor screening. J. Infect. Dis. 2010, 202, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Chancey, C.; Grinev, A.; Volkova, E.; Rios, M. The global ecology and epidemiology of West Nile virus. Biomed. Res. Int. 2015, 2015, 376230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linke, S.; Ellerbrok, H.; Niedrig, M.; Nitsche, A.; Pauli, G. Detection of West Nile virus lineages 1 and 2 by real-time PCR. J. Virol. Methods 2007, 146, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; Terenius, O.; Oshaghi, M.A.; Motazakker, M.; Asgari, S.; Dabiri, F.; Vatandoost, H.; Mohammadi Bavani, M.; Chavshin, A.R. West Nile virus in mosquitoes of Iranian wetlands. Vector Borne Zoonotic Dis. 2015, 15, 750–754. [Google Scholar] [CrossRef]
- Pesko, K.N.; Ebel, G.D. West Nile virus population genetics and evolution. Infect. Genet. Evol. 2012, 12, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Shah-Hosseini, N.; Chinikar, S.; Ataei, B.; Fooks, A.R.; Groschup, M.H. Phylogenetic analysis of West Nile virus genome, Iran. Emerg. Infect. Dis. 2014, 20, 1419–1421. [Google Scholar] [CrossRef] [Green Version]
- Shahhosseini, N.; Chinikar, S.; Moosa-Kazemi, S.H.; Sedaghat, M.M.; Kayedi, M.H.; Lühken, R.; Schmidt-Chanasit, J. West Nile Virus lineage-2 in culex specimens from Iran. Trop. Med. Int. Health 2017, 22, 1343–1349. [Google Scholar] [CrossRef] [Green Version]
- Venter, M.; Human, S.; Zaayman, D.; Gerdes, G.H.; Williams, J.; Steyl, J.; Leman, P.A.; Paweska, J.T.; Setzkorn, H.; Rous, G. Lineage 2 West Nile virus as cause of fatal neurologic disease in horses, South Africa. Emerg. Infect. Dis. 2009, 15, 877–884. [Google Scholar] [CrossRef]
- Danis, K.; Papa, A.; Theocharopoulos, G.; Dougas, G.; Athanasiou, M.; Detsis, M.; Baka, A.; Lytras, T.; Mellou, K.; Bonovas, S. Outbreak of West Nile virus infection in Greece, 2010. Emerg. Infect. Dis. 2011, 17, 1868–1872. [Google Scholar] [CrossRef]
- Papa, A.; Papadopoulou, E.; Chatzixanthouliou, C.; Glouftsios, P.; Pappa, S.; Pervanidou, D.; Georgiou, L. Emergence of West Nile virus lineage 2 belonging to the Eastern European subclade, Greece. Arch. Virol. 2019, 164, 1673–1675. [Google Scholar] [CrossRef]
- Veo, C.; Della Ventura, C.; Moreno, A.; Rovida, F.; Percivalle, E.; Canziani, S.; Torri, D.; Calzolari, M.; Baldanti, F.; Galli, M. Evolutionary dynamics of the lineage 2 West Nile virus that caused the largest European epidemic: Italy 2011–2018. Viruses 2019, 11, 814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakonyi, T.; Hubálek, Z.; Rudolf, I.; Nowotny, N. Novel flavivirus or new lineage of West Nile virus, central Europe. Emerg. Infect. Dis. 2005, 11, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Campbell, G.L.; Marfin, A.A.; Lanciotti, R.S.; Gubler, D.J. West Nile virus. Lancet Infect. Dis. 2002, 2, 519–529. [Google Scholar] [CrossRef]
- Eybpoosh, S.; Fazlalipour, M.; Baniasadi, V.; Pouriayevali, M.H.; Sadeghi, F.; Vasmehjani, A.A.; Niya, M.H.K.; Hewson, R.; Salehi-Vaziri, M. Epidemiology of West Nile Virus in the Eastern Mediterranean region: A systematic review. PLoS Negl. Trop. Dis. 2019, 13, e0007081. [Google Scholar] [CrossRef]
- Ahmadnejad, F.; Otarod, V.; Fallah, M.; Lowenski, S.; Sedighi-Moghaddam, R.; Zavareh, A.; Durand, B.; Lecollinet, S.; Sabatier, P. Spread of West Nile virus in Iran: A cross-sectional serosurvey in equines, 2008–2009. Epidemiol. Infect. 2011, 139, 1587–1593. [Google Scholar] [CrossRef]
- Chinikar, S.; Javadi, A.; Ataei, B.; Shakeri, H.; Moradi, M.; Mostafavi, E.; Ghiasi, S.M. Detection of West Nile virus genome and specific antibodies in Iranian encephalitis patients. Epidemiol. Infect. 2012, 140, 1525–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinikar, S.; Shah-Hosseini, N.; Mostafavi, E.; Moradi, M.; Khakifirouz, S.; Jalali, T.; Goya, M.M.; Shirzadi, M.R.; Zainali, M.; Fooks, A.R. Seroprevalence of West Nile virus in Iran. Vector Borne Zoonotic Dis. 2013, 13, 586–589. [Google Scholar] [CrossRef]
- Fereidouni, S.R.; Ziegler, U.; Linke, S.; Niedrig, M.; Modirrousta, H.; Hoffmann, B.; Groschup, M.H. West Nile virus monitoring in migrating and resident water birds in Iran: Are common coots the main reservoirs of the virus in wetlands? Vector Borne Zoonotic Dis. 2011, 11, 1377–1381. [Google Scholar] [CrossRef]
- Sharifi, Z.; Mahmoudian, S.M.; Talebian, A. A study of West Nile virus infection in Iranian blood donors. Arch. Iran. Med. 2010, 13, 1–4. [Google Scholar]
- Zohaib, A.; Niazi, S.K.; Saqib, M.; Sajid, M.S.; Khan, I.; Athar, M.A.; Taj, Z.; Abbas, G.; Rathore, M.A.; Ghani, E. Detection of West Nile virus lineage 1 sequences in blood donors, Punjab Province, Pakistan. Int. J. Infect. Dis. 2019, 81, 137–139. [Google Scholar] [CrossRef] [Green Version]
- Akıner, M.M.; Öztürk, M.; Başer, A.B.; Günay, F.; Hacıoğlu, S.; Brinkmann, A.; Emanet, N.; Alten, B.; Özkul, A.; Nitsche, A. Arboviral screening of invasive Aedes species in northeastern Turkey: West Nile virus circulation and detection of insect-only viruses. PLoS Negl. Trop. Dis. 2019, 13, e0007334. [Google Scholar] [CrossRef] [PubMed]
- Ergunay, K.; Gunay, F.; Kasap, O.E.; Oter, K.; Gargari, S.; Karaoglu, T.; Tezcan, S.; Cabalar, M.; Yildirim, Y.; Emekdas, G. Serological, molecular and entomological surveillance demonstrates widespread circulation of West Nile virus in Turkey. PLoS Negl. Trop. Dis. 2014, 8, e3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozkul, A.; Ergunay, K.; Koysuren, A.; Alkan, F.; Arsava, E.M.; Tezcan, S.; Emekdas, G.; Hacioglu, S.; Turan, M.; Us, D. Concurrent occurrence of human and equine West Nile virus infections in Central Anatolia, Turkey: The first evidence for circulation of lineage 1 viruses. Int. J. Infect. Dis. 2013, 17, e546–e551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocal, M.; Orsten, S.; Inkaya, A.; Yetim, E.; Acar, N.; Alp, S.; Erisoz Kasap, O.; Gunay, F.; Arsava, E.; Alten, B. Ongoing Activity of Toscana Virus Genotype A and West Nile Virus Lineage 1 Strains in T urkey: A Clinical and Field Survey. Zoonoses Public Health 2014, 61, 480–491. [Google Scholar] [CrossRef]
- Lambrechts, L.; Chevillon, C.; Albright, R.G.; Thaisomboonsuk, B.; Richardson, J.H.; Jarman, R.G.; Scott, T.W. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC. Evol. Biol. 2009, 9, 160. [Google Scholar] [CrossRef] [Green Version]
- Brustolin, M.; Talavera, S.; Santamaría, C.; Rivas, R.; Pujol, N.; Aranda, C.; Marquès, E.; Valle, M.; Verdún, M.; Pagès, N. Culex pipiens and Stegomyia albopicta (= Aedes albopictus) populations as vectors for lineage 1 and 2 West Nile virus in Europe. Med. Vet. Entomol. 2016, 30, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Azari-Hamidian, S.; Norouzi, B.; Harbach, R.E. A detailed review of the mosquitoes (Diptera: Culicidae) of Iran and their medical and veterinary importance. Acta Trop. 2019, 194, 106–122. [Google Scholar] [CrossRef]
- Azari-Hamidian, S.; Harbach, R.E. Keys to the adult females and fourth-instar larvae of the mosquitoes of Iran (Diptera: Culicidae). Zootaxa 2009, 2078, 1–33. [Google Scholar] [CrossRef]
- Vazeille-Falcoz, M.; Mousson, L.; Rodhain, F.; Chungue, E.; Failloux, A.-B. Variation in oral susceptibility to dengue type 2 virus of populations of Aedes aegypti from the islands of Tahiti and Moorea, French Polynesia. Am. J. Trop. Med. Hyg. 1999, 60, 292–299. [Google Scholar] [CrossRef] [Green Version]
- Zakhia, R.; Mousson, L.; Vazeille, M.; Haddad, N.; Failloux, A.-B. Experimental transmission of West Nile Virus and Rift Valley Fever Virus by Culex pipiens from Lebanon. PLoS Negl. Trop. Dis. 2018, 12, e0005983. [Google Scholar] [CrossRef]
- Murgue, B.; Murri, S.; Zientara, S.; Durand, B.; Durand, J.-P.; Zeller, H. West Nile outbreak in horses in southern France, 2000: The return after 35 years. Emerg. Infect. Dis. 2001, 7, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Dubrulle, M.; Mousson, L.; Moutailler, S.; Vazeille, M.; Failloux, A.-B. Chikungunya virus and Aedes mosquitoes: Saliva is infectious as soon as two days after oral infection. PLoS ONE 2009, 4, e5895. [Google Scholar] [CrossRef] [PubMed]
- Jupille, H.; Seixas, G.; Mousson, L.; Sousa, C.A.; Failloux, A.-B. Zika virus, a new threat for Europe? PLoS Negl. Trop. Dis. 2016, 10, e0004901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chouin-Carneiro, T.; Vega-Rua, A.; Vazeille, M.; Yebakima, A.; Girod, R.; Goindin, D.; Dupont-Rouzeyrol, M.; Lourenco-de-Oliveira, R.; Failloux, A.-B. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLoS Negl. Trop. Dis. 2016, 10, e0004543. [Google Scholar] [CrossRef]
- Amraoui, F.; Krida, G.; Bouattour, A.; Rhim, A.; Daaboub, J.; Harrat, Z.; Boubidi, S.-C.; Tijane, M.; Sarih, M.; Failloux, A.-B. Culex pipiens, an experimental efficient vector of West Nile and Rift Valley fever viruses in the Maghreb region. PLoS ONE 2012, 7, e36757. [Google Scholar] [CrossRef] [Green Version]
- Balenghien, T.; Vazeille, M.; Grandadam, M.; Schaffner, F.; Zeller, H.; Reiter, P.; Sabatier, P.; Fouque, F.; Bicout, D.J. Vector competence of some French Culex and Aedes mosquitoes for West Nile virus. Vector Borne Zoonotic Dis. 2008, 8, 589–596. [Google Scholar] [CrossRef]
- Balenghien, T.; Vazeille, M.; Reiter, P.; Schaffner, F.; Zeller, H.; Bicout, D.J. Evidence of laboratory vector competence of Culex modestus for West Nile virus. J. Am. Mosq. Control. Assoc. 2007, 23, 233–237. [Google Scholar] [CrossRef]
- Ebel, G.D.; Rochlin, I.; Longacker, J.; Kramer, L.D. Culex restuans (Diptera: Culicidae) relative abundance and vector competence for West Nile Virus. J. Med. Entomol. 2005, 42, 838–843. [Google Scholar] [CrossRef]
- Goddard, L.B.; Roth, A.E.; Reisen, W.K.; Scott, T.W. Vector competence of California mosquitoes for West Nile virus. Emerg. Infect. Dis. 2002, 8, 1385–1391. [Google Scholar] [CrossRef]
- Sardelis, M.R.; Turell, M.J.; Dohm, D.J.; O’Guinn, M.L. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg. Infect. Dis. 2001, 7, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Turell, M.J.; Dohm, D.J.; Sardelis, M.R.; O’guinn, M.L.; Andreadis, T.G.; Blow, J.A. An update on the potential of North American mosquitoes (Diptera: Culicidae) to transmit West Nile virus. J. Med. Entomol. 2005, 42, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Turell, M.J.; O’Guinn, M.L.; Dohm, D.J.; Webb, J.P., Jr.; Sardelis, M.R. Vector competence of Culex tarsalis from orange County, California, for West Nile virus. Vector Borne Zoonotic Dis. 2002, 2, 193–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogels, C.; Göertz, G.; Pijlman, G.; Koenraadt, C. Vector competence of northern and southern European Culex pipiens pipiens mosquitoes for West Nile virus across a gradient of temperatures. Med. Vet. Entomol. 2017, 31, 358–364. [Google Scholar] [CrossRef]
- Flores, F.S.; Zanluca, C.; Guglielmone, A.A.; Duarte Dos Santos, C.N.; Labruna, M.B.; Diaz, A. Vector Competence for West Nile Virus and St. Louis Encephalitis Virus (Flavivirus) of Three Tick Species of the Genus Amblyomma (Acari: Ixodidae). Am. J. Trop. Med. Hyg. 2019, 100, 1230–1235. [Google Scholar] [CrossRef] [Green Version]
- Zaim, M. The distribution and larval habitat characteristics of Iranian Culicinae. J. Am. Mosq. Control. Assoc. 1987, 3, 568–573. [Google Scholar]
- Ciota, A.T.; Kramer, L.D. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 2013, 5, 3021–3047. [Google Scholar] [CrossRef]
- Hutcheson, H.J.; Gorham, C.H.; Machain-Williams, C.; Loroño-Pino, M.A.; James, A.M.; Marlenee, N.L.; Winn, B.; Beaty, B.J.; Blair, C.D. Experimental transmission of West Nile virus (Flaviviridae: Flavivirus) by Carios capensis ticks from North America. Vector Borne Zoonotic Dis. 2005, 5, 293–295. [Google Scholar] [CrossRef]
- Moro, C.V.; Chauve, C.; Zenner, L. Vectorial role of some dermanyssoid mites (Acari, Mesostigmata, Dermanyssoidea). Parasite 2005, 12, 99–109. [Google Scholar] [CrossRef]
- Tempelis, C.H. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J. Med. Entomol. 1975, 11, 635–653. [Google Scholar] [CrossRef]
- Kramer, L.D.; Ebel, G.D. Dynamics of flavivirus infection in mosquitoes. Adv. Viruses Res. 2003, 60, 187–232. [Google Scholar]
- Anderson, S.L.; Richards, S.L.; Tabachnick, W.J.; Smartt, C.T. Effects of West Nile virus dose and extrinsic incubation temperature on temporal progression of vector competence in Culex pipiens quinquefasciatus. J. Am. Mosq. Control. Assoc. 2010, 26, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Paz, S. Effects of climate change on vector-borne diseases: An updated focus on West Nile virus in humans. Emerg. Top. Life Sci. 2019, 3, 143–152. [Google Scholar]
- Turell, M.; O’Guinn, M.; Oliver, J. Potential for New York mosquitoes to transmit West Nile virus. Am. J. Trop. Med. Hyg. 2000, 62, 413–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fansiri, T.; Fontaine, A.; Diancourt, L.; Caro, V.; Thaisomboonsuk, B.; Richardson, J.H.; Jarman, R.G.; Ponlawat, A.; Lambrechts, L. Genetic mapping of specific interactions between Aedes aegypti mosquitoes and dengue viruses. PLoS Genet. 2013, 9, e1003621. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakhshi, H.; Mousson, L.; Vazeille, M.; Zakeri, S.; Raz, A.; de Lamballerie, X.; Dinparast-Djadid, N.; Failloux, A.-B. High Transmission Potential of West Nile Virus Lineage 1 for Cx. pipiens s.l. of Iran. Viruses 2020, 12, 397. https://doi.org/10.3390/v12040397
Bakhshi H, Mousson L, Vazeille M, Zakeri S, Raz A, de Lamballerie X, Dinparast-Djadid N, Failloux A-B. High Transmission Potential of West Nile Virus Lineage 1 for Cx. pipiens s.l. of Iran. Viruses. 2020; 12(4):397. https://doi.org/10.3390/v12040397
Chicago/Turabian StyleBakhshi, Hasan, Laurence Mousson, Marie Vazeille, Sedigheh Zakeri, Abbasali Raz, Xavier de Lamballerie, Navid Dinparast-Djadid, and Anna-Bella Failloux. 2020. "High Transmission Potential of West Nile Virus Lineage 1 for Cx. pipiens s.l. of Iran" Viruses 12, no. 4: 397. https://doi.org/10.3390/v12040397
APA StyleBakhshi, H., Mousson, L., Vazeille, M., Zakeri, S., Raz, A., de Lamballerie, X., Dinparast-Djadid, N., & Failloux, A.-B. (2020). High Transmission Potential of West Nile Virus Lineage 1 for Cx. pipiens s.l. of Iran. Viruses, 12(4), 397. https://doi.org/10.3390/v12040397





