RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Used in the Study
2.2. RNA Isolation, Library Preparation and Sequencing
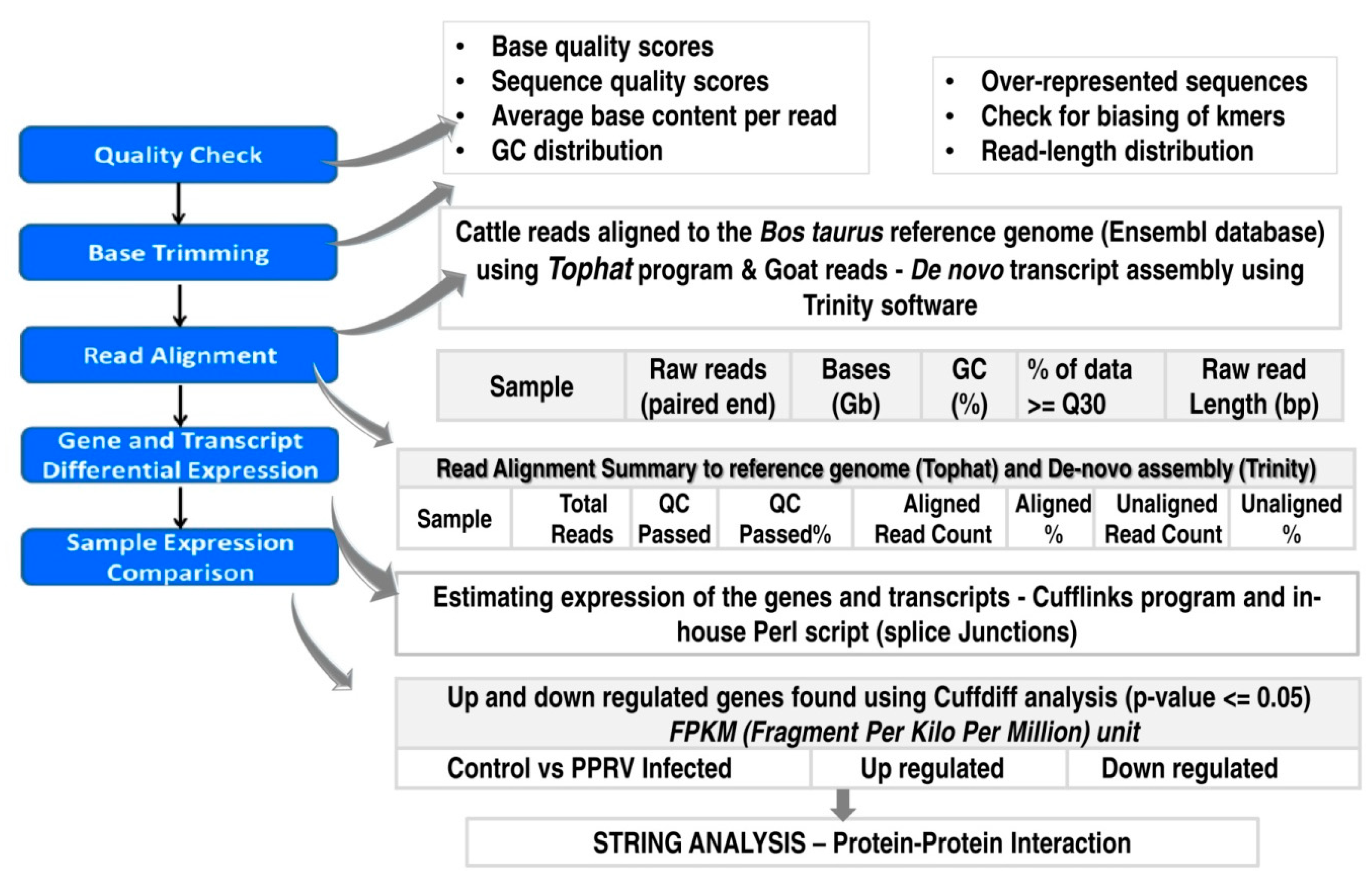
2.3. Bioinformatic Analysis of the Sequence Data
2.3.1. Quality Check and Assembly
2.3.2. Transcriptome Annotation, Differential Gene Expression Analysis and Protein-Protein Interactions
2.3.3. Validation of the Differential Targets Identified in RNAseq Data
3. Results
3.1. Transcriptome Assembly, Annotation and Gene Ontology
3.1.1. Transcriptome Assembly
3.1.2. Annotation:
3.1.3. Gene Ontology
3.2. Functional Analysis of Differentially Expressed Genes across Goat and Cattle upon Exposure to PPRV
3.3. PPRV Infection Induces Different Immune Responses in Goats and Cattle
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lefevre, P.C.; Diallo, A. Peste des petits ruminants. Rev. Sci. Tech. Off. Int. Epiz. 1990, 9, 951–965. [Google Scholar] [CrossRef]
- Shaila, M.S.; Purushothaman, V.; Bhavasar, D.; Venugopal, K.; Venkatesan, R.A. Peste des petits ruminants of sheep in India. Vet. Rec. 1989, 125, 602. [Google Scholar] [PubMed]
- Kulkarni, D.D.; Bhikane, A.U.; Shaila, M.S.; Varalakshmi, P.; Apte, M.P.; Narladkar, B.W. Peste des petits ruminants in goats in India. Vet. Rec. 1996, 138, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Nanda, Y.P.; Chatterjee, A.; Purohit, A.K.; Diallo, A.; Innui, K.; Sharma, R.N.; Libeau, G.; Thevasagayam, J.A.; Burning, A.; Kitching, R.P.; et al. The isolation of Peste des petitsruminants virus from Northern India. Vet. Microbiol. 1996, 51, 207–212. [Google Scholar] [CrossRef]
- Shaila, M.S.; Peter, A.B.; Varalakshmi, P.; Apte, M.P.; Rajendran, M.P.; Anbumani, S.P. Peste des petits ruminants in Tamilnadu goats. Indian Vet. J. 1996, 73, 587–588. [Google Scholar]
- Saravanan, P.; Sen, A.; Balamurugan, V.; Rajak, K.K.; Bhanuprakash, V.; Palaniswami, K.S.; Nachimuthu, K.; Thangavelu, A.; Dhinakarraj, G.; Hegde, R.; et al. Comparative efficacy of Peste des petits ruminants (PPR) vaccines. Biologicals 2010, 38, 479–485. [Google Scholar] [CrossRef]
- Couacy-Hymann, E.; Bodjo, C.; Danho, T.; Diallo, G.L.A. Evaluation of the virulence of some strains of Peste-des-petits-ruminants virus (PPRV) in experimentally infected West African dwarf goats. Vet. J. 2007, 173, 178–183. [Google Scholar] [CrossRef]
- Singh, R.P.; Saravanan, B.P.; Dhar, P.; Shah, L.C.; Bandyopadhyay, S.K. Development of a monoclonal antibody based competitive-ELISA for detection and titration of antibodies to Peste des petites ruminants (PPR) virus. Vet. Microbiol. 2004, 98, 3–15. [Google Scholar] [CrossRef]
- Delil, F.; Asfaw, Y.; Gebreegziabher, B. Prevalence of antibodies to Peste des petits ruminants virus before and during outbreaks of the disease in Awash Fentale district; Afar; Ethiopia. Trop. Anim. Health Prod. 2012, 44, 1329–1330. [Google Scholar] [CrossRef]
- Anderson, J.; McKay, A. The detection of antibodies against Peste des petits ruminants virus in cattle; sheep and goats and the possible implications to rinderpest control programmes. Epidemiol. Infect. 1994, 112, 225–231. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.A.; Siddique, M.; Sajjad, U.R.; Abubakar, M.; Ashraf, M. The detection of antibody against Peste des petits ruminants virus in sheep; goats; cattle and buffaloes. Trop. Anim. Health Prod. 2008, 40, 521–527. [Google Scholar] [CrossRef]
- Mornet, P.; Orue, J.; Gillbert, Y.; Thiery, G.S.M. La peste des petits Ruminants en Afrique occidentale française ses rapports avec la Peste Bovine. Rev. Elev. Med. Vet. Pays Trop. 1956, 9, 313–342. [Google Scholar] [CrossRef] [Green Version]
- Govindarajan, R.; Koteeswaran, A.; Venugopalan, A.T.; Shyam, G.; Shaouna, S.; Shail, M.S.; Ramachandran, S. Isolation of Pestes des petits ruminants virus from an outbreak in Indian buffalo (Bubalus bubalis). Vet. Rec. 1997, 141, 573–574. [Google Scholar] [CrossRef]
- Roger, F.; Guebre, Y.M.; Libeau, G.; Diallo, A.; Yigezu, L.M.; Yilma, T. Detection of antibodies of rinderpest and Peste des petits ruminants viruses (Paramyxoviridae; Morbillivirus) during a new epizootic disease in Ethiopian camels (Camelusdromedarius). Rev. Med. Vet. 2001, 152, 265–268. [Google Scholar]
- Pawar, R.M.; Raj, G.D.; Balachandran, C. Relationship between the level of signaling lymphocyte activation molecule mRNA and replication of Peste des petits ruminants virus in peripheral blood mononuclear cells of host animals. ActaVirol. 2008, 52, 231–236. [Google Scholar]
- Birch, J.; Juleff, N.; Heaton, M.P.; Kalbfleisch, T.; Kijas, J.; Bailey, D. Characterization of ovine nectin-4; a novel Peste des petits ruminants virus receptor. J. Virol. 2013, 87, 4756–4761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhanasekaran, S.; Biswas, M.; Ambothi, R.; Ramya, V.R.; Raj, G.D.; Tirumurugaan, K.G.; Raja, A.; Kataria, R.S.; Parida, S.; Subbiah, E. Toll-Like Receptor Responses to Peste des petits ruminants Virus in Goats and Water Buffalo. PLoS ONE 2014, 9, 1609–1611. [Google Scholar] [CrossRef]
- Baron, M.D.; Diallo, A.; Lancelot, R.; Libeau, G. Chapter One—Peste des Petits Ruminants Virus. In Advances in Virus Research; Margaret Kielian, K.M., Thomas, C.M., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 95, pp. 1–42. [Google Scholar]
- Ozkul, A.; Akca, Y.; Alkan, F.; Barrett, T.; Karaoglu, T.; Daglap, S.B.; Anderson, J.; Yesilbag, K.; Cokcaliskan, C.; Genacy, A.; et al. Prevalence; distribution and host range of peste des petitsruminants virus; Turkey. Emerg. Infect. Dis. 2002, 8, 708–712. [Google Scholar] [CrossRef]
- Abraham, G. Antibody seroprevalences against Peste des petitsruminants virus in camels; cattle; goats and sheep in Ethiopia. Prev. Vet. Med. 2005, 70, 51–57. [Google Scholar] [CrossRef]
- Khalafalla, A.I.; Saeed, I.K.; Ali, Y.H.; Abdurrahman, M.B.; Kwiatek, O.; Libeau, G.; Obeida, A.A.; Abbas, Z. An outbreak of peste des petits ruminants (PPR) in camels in the Sudan. Acta Trop. 2010, 116, 161–165. [Google Scholar] [CrossRef]
- Sharon, D.; Chenb, R.; Snyderb, M. Systems biology approaches to disease marker discovery. Dis. Markers 2010, 28, 209–224. [Google Scholar] [CrossRef]
- Morrison-Smith, S.; Boucher, C.; Bunt, A.; Ruiz, J. Elucidating the role and use of bioinformatics software in life science research. In Proceedings of the 2015 British HCI Conference, Lincoln, UK, 13–17 July 2015; pp. 230–238. [Google Scholar]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses, an interplay between induction; signalling; antiviral responses and virus counter measures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- Atmaca, H.T.; Kul, O. Examination of epithelial tissue cytokine response to natural Peste des petits ruminants virus (PPRV) infection in sheep and goats by immunohistochemistry. Histol. Histopathol. 2012, 27, 69–78. [Google Scholar]
- Baron, J.; Bin-Tarif, A.; Herbert, R.; Frost, L.; Taylor, G.; Baron, M.D. Early changes in cytokine expression in Peste des petits ruminants disease. Vet. Res. 2014, 45, 22. [Google Scholar] [CrossRef] [Green Version]
- Fontana, J.M.; Bankamp, B.; Bellini, W.J.; Rota, P.A. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 2008, 374, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Caignard, G.; Bourai, M.; Jacob, Y.; Tangy, F.; Vidalain, P.O. Inhibition of IFN-alpha/ beta signalling by two discrete peptides within measles virus V protein that specifically bind STAT1 and STAT2. Virology 2009, 383, 112–120. [Google Scholar] [CrossRef]
- Manjunath, S.; Kumar, G.R.; Mishra, B.P.; Mishra, B.; Sahoo, A.P.; Joshi, C.G.; Tiwari, A.K.; Rajak, K.K.; Janga, S.C. Genomic analysis of host- Peste des petits ruminants vaccine viral transcriptome uncovers transcription factors modulating immune regulatory pathways. Vet. Res. 2015, 46, 15. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Yang, X.; Nian, X.; Zhang, Z.; Dou, Y.; Zhang, X.; Luo, X.; Su, J.; Zhu, Q.; Cai, X. Identification of amino-acid residues in the V protein of peste des petits ruminants essential for interference and suppression of STAT-mediated interferon signaling. Virology 2015, 483, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Wang, T.; Xue, Q.; Li, Z.; Yang, B.; Wang, J. MicroRNA expression profiling of goat peripheral blood mononuclear cells in response to Peste des petits ruminants virus infection. Vet. Res. 2018, 49, 62. [Google Scholar] [CrossRef] [Green Version]
- Wani, S.A.; Sahu, A.R.; Khan, R.I.N.; Pandey, A.; Saxena, S.; Hosamani, N.; Malla, W.A.; Chaudhary, D.; Kanchan, S.; Sah, V.; et al. Contrasting Gene Expression Profiles of Monocytes and Lymphocytes From Peste des petits ruminants Virus Infected Goats. Front. Immunol. 2019, 10, 1463. [Google Scholar] [CrossRef]
- De Veer, M.J.; Holko, M.; Frevel, M.; Walker, E.; Der, S.; Paranjape, J.M.; Silverman, R.H.; Williams, B.R.G. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001, 69, 912–920. [Google Scholar]
- Sarasin-Filipowicz, M.; Oakeley, E.J.; Duong, F.H.T.; Christen, V.; Terracciano, L.; Filipowicz, W.; Heim, M.H. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA 2008, 105, 7034–7039. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Eisenächer, K.; Kirchhofer, A.; Brzózka, K.; Lammens, A.; Lammens, K.; Fujita, T.; Conzelmann, K.-K.; Krug, A.B.; Hopfner, K.-P. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol. Cell. 2008, 29, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, P.S.; Jatta, K.; Wågsäter, D.; Gredmark, S.; Hedin, U.; Paulsson-Berne, G.; Söderberg-Nauclér, C.; Hansson, G.K.; Sirsjö, A. The antiviral cytomegalovirus inducible gene 5/viperin is expressed in atherosclerosis and regulated by pro-inflammatory agents. Arterioscler. Thromb. Vasc. Biol. 2005, 25, e113–e116. [Google Scholar] [CrossRef]
- Severa, M.; Coccia, E.M.; Fitzgerald, K.A. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 2006, 281, 26188–26195. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Guo, H.; Xu, C.; Chang, J.; Gu, B.; Wang, L.; Block, T.M.; Guo, J.-T. Identification of Three Interferon-Inducible Cellular Enzymes That Inhibit the Replication of Hepatitis C Virus. J. Virol. 2008, 82, 1665–1678. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Weidner, J.M.; Qing, M.; Pan, X.B.; Guo, H.; Xu, C.; Zhang, X.; Birk, A.; Chang, J.; Shi, P.Y.; et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 2010, 84, 8332–8341. [Google Scholar] [CrossRef] [Green Version]
- Rivieccio, M.A.; Suh, H.S.; Zhao, Y.; Zhao, M.-L.; Chin, K.C.; Lee, S.C.; Brosnan, C.F. TLR3 ligation activates an antiviral response in human fetal astrocytes, a role for viperin/ cig5. J. Immunol. 2006, 177, 4735–4741. [Google Scholar] [CrossRef] [Green Version]
- Stirnweiss, A.; Ksienzyk, A.; Klages, K.; Rand, U.; Grashoff, M.; Hauser, H.; Kröger, A. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 2010, 184, 5179–5185. [Google Scholar] [CrossRef]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Gene Target | Sequence (5′----->3′) | Nucleotide Position | Product Size |
|---|---|---|---|
| PPRV-M-F | AGTGATTGAGGATAACGAC | 3608–3627 | 242 bp |
| PPRV-M-R | TTAGCGCTAAACACACTTCC | 3850–3830 | |
| β-actin F | GGCTGTGCTGTCCCTGTAC | 156–174 | 156 bp |
| β-actin R | CCGGAGTCCATCACGATGC | 214–194 | |
| GAPDHF | GGCGTGAACCACGAGAAGTATAA | 470–492 | 119 bp |
| GAPDHR | CCCTCCACGATGCCAAAGT | 588–570 | |
| DHX58F | GGGACATGCTGAAGAAGCTC | 723–742 | 175 bp |
| DHX58R | TTGTACCGACGCAGATGAAG | 897–878 | |
| IFI44F | GTCCATGTGGTGTTGCTCAC | 1157–1176 | 180 bp |
| IFI44R | CTCCAGTTCCCACTCTGAGG | 1336–1317 | |
| IFI6F | AACTCGT/GTGGCCTCCTCAC | 524–542 | 168 bp |
| IFI6R | TTCGACTTGCTTGTGGACAG | 691–672 | |
| ISG15F | CAGTTCATCGCCCAGAAGAT | 147–166 | 171 bp |
| ISG15R | GTCGTTCCTCACCAGGATGT | 317–298 | |
| IFIL1F | ACTACGGCCGATTTCTGGAA | 1467–1485 | 244 bp |
| IFIL1R | AGGGCCCGCTCATAATACTC | 1709–1690 | |
| IFIL2F | TGCTGCTATAGGGCCAAAGT | 1084–1103 | 240 bp |
| IFIL2R | CACTTGTTTGGCTACG/AGGAG | 1323–1304 | |
| IFIL3F | GAGCTGGACTGTGAGGAAGG | 461–480 | 242 bp |
| IFIL3R | AGGGCCAGGAGAACTTTGAT | 702–683 | |
| MX1F | GTCCCTGCTAACGTGGACAT | 714–733 | 155 bp |
| MX1R | ACCAGGTTTCTCACCACGTC | 868–849 | |
| MX2F | GCAGATCAAGGCACTCATCA | 992–1011 | 168 bp |
| MX2R | ACCAGGTCTGGTTTGGTCAG | 1159–1140 | |
| OAS1XF | CTGACCTCGTCGTCTTCCTC | 260–279 | 224 |
| OAS1XR | CAGGACATCGAACTCCACCT | 483–464 | |
| PDE12F | GTGTTTCGAATCAAGCAGCA | 1186–1205 | 176 bp |
| PDE12R | CTCTGGAGCACCCTTTCTTG | 1361–1342 | |
| RSAD2F | ATTTGGACATTCTCGCCATC | 570–589 | 182 bp |
| RSAD2 R | ATCCTCGTCCACGTTGAATC | 751–732 | |
| DDX58F | AAGGAT/CTGCCTCCATGACTG | 2747–2766 | 164 bp |
| DDX58R | GCAGCATCAAATGGGATCTT | 2910–2891 | |
| IFI44LF | TTCCAAGGCCATTTGACTCG | 798–817 | 150 bp |
| IFI44LR | CTCTTT/CCTCATCCAGCCCCA | 947–928 | |
| IFIH1F | ATCTCGTGTTTCAGGGCCAG | 389–408 | 206 bp |
| IFIH1R | AGGGCCTCTACGAACATCCT | 594–575 | |
| IRF7F | TCCCCGCACTACACCATCTA | 1614–1633 | 224 bp |
| IRF7R | AAGTGCTCCAGGAAGTGCTC | 1837–1818 |
| Sample/SRA BioSample ID/SRA ID | No. of Paired End Reads and No. of Bases (Gb) | Aligned Read Count and Percent Aligned | % Q > 30 | Number of Transcripts with FPKM ≥ 1.0 |
|---|---|---|---|---|
| NGS data generated from the Project–P4365 | ||||
| Goat_4-Control: [BioSample ID–SAMN14478390 & SRA ID–SRR11440511] | 23,662,652 (4.73 Gb) | 14,740,174 (86.83%) | 90.62 | 179,233 ** |
| Goat_3- PPRV Infected: [BioSample ID–SAMN14469261 & SRA ID–SRR11434618] | 30,098,206 (6.01 Gb) | 18,537,737 (87.77%) | 91.36 | 150,093 ** |
| Cattle_4- Control: [BioSample ID–SAMN14480722 & SRA ID–SRR11442915] | 25,656,128 (5.131 Gb) | 44,579,027 (90.11%) | 91.3 | 11503 |
| Cattle_3- PPRVInfected: [BioSample ID–SAMN14480403 & SRA ID–SRR11442832] | 25,995,419 (5.199 Gb) | 36,088,556 (71.61%) | 91.25 | 11633 |
| NGS data generated from the Project–P4117 | ||||
| Goat_2-Control: [BioSample ID–SAMN14492330 & SRA ID–SRR11431701] | 18,315,355 (3.66 Gb) | 13,017,046 (89.76%) | 87.95 | 83,315 ** |
| Goat_1-PPRV Infected: [BioSample ID–SAMN14466456 & SRA ID–SRR11431292] | 27,803,932 (3.54 Gb) | 17,955,944 (88.27%) | 87.87 | 71,874 ** |
| Cattle_2-Control: [BioSample ID–SAMN14480879 & SRA ID–SRR11443109] | 24,457,636 (3.08 Gb) | 19,605,498 (63.68%) | 87.54 | 10,414 |
| Cattle_1-PPRV infected: [BioSample ID–SAMN14478687 & SRA ID–SRR11440572] | 25,055,747 (3.23 Gb) | 14,281,792 (57.00%) | 89.64 | 10,356 |
| FPKM | Number of Transcripts (P4365) * | Number of Transcripts (P4117) * | ||||||
|---|---|---|---|---|---|---|---|---|
| Goat_4 Cont. ** | Goat_3 PPRV Inf. ** | Cattle_4Cont. | Cattle_3 PRV Inf. | Goat_2 Cont. ** | Goat_1 PPRV Inf. ** | Cattle_2Cont. | Cattle_1PRV Inf. | |
| 1–2 | 89,644 | 73,363 | 959 | 1048 | 44,664 | 32,444 | 1128 | 1111 |
| 2–5 | 46,460 | 37,026 | 1753 | 1706 | 16,919 | 17,975 | 2238 | 2287 |
| 5–10 | 17,944 | 17,794 | 1933 | 1914 | 9644 | 9809 | 2267 | 2215 |
| 10–20 | 14,126 | 12,592 | 2338 | 2398 | 6500 | 6250 | 2160 | 2000 |
| 20–50 | 8425 | 6863 | 2723 | 2700 | 3775 | 3522 | 1668 | 1611 |
| 50–100 | 1665 | 1524 | 978 | 1013 | 992 | 1009 | 473 | 561 |
| ≥100 | 969 | 931 | 819 | 854 | 821 | 865 | 480 | 571 |
| Total | 179,233 | 150,093 | 11,503 | 11,633 | 83,315 | 71,874 | 10,414 | 10,356 |
| 3,52,462 assembled transcripts (longest transcript 23,144 bases) | 1,75,959 assembled transcripts (longest transcript 30,976 bases) | |||||||
| Deseq Combination | Down-Regulated Transcripts | Up-Regulated Transcripts |
|---|---|---|
| Goat Cont. vs. Goat Inf. | 474 (19.0%) | 2017 (81.0%) |
| Cattle Cont. vs. Cattle Inf. | 368 (68.0%) | 173 (32.0%) |
| Goat Cont. vs. Cattle Cont. | 1574 (60.7%) | 1018 (39.3%) |
| S.No | Gene ID | Gene | Goat Control FPKM Value | Goat Infected FPKM Value | log2 (Fold Change) |
|---|---|---|---|---|---|
| 1 | ENSBTAG00000007554 | IFI6 | 685.79 | 2507.99 | 1.87 |
| 2 | ENSBTAG00000007881 | IFIT1 | 66.63 | 247.95 | 1.90 |
| 3 | ENSBTAG00000008471 | MX2 | 75.59 | 235.75 | 1.64 |
| 4 | ENSBTAG00000009768 | IFIT3 | 116.90 | 403.79 | 1.79 |
| 5 | ENSBTAG00000014707 | ISG15 | 725.86 | 2915.81 | 2.01 |
| 6 | ENSBTAG00000016061 | RSAD2 | 74.83 | 439.60 | 2.55 |
| 7 | ENSBTAG00000030913 | Mx1 | 266.28 | 1029.88 | 1.95 |
| 8 | ENSBTAG00000034349 | IFI44 | 41.14 | 151.84 | 1.88 |
| 9 | ENSBTAG00000034918 | IFIT2 | 144.59 | 588.48 | 2.03 |
| 10 | ENSBTAG00000037527 | OAS1X | 60.86 | 258.11 | 2.08 |
| 11 | ENSBTAG00000046580 | DHX58 | 74.18 | 224.04 | 1.59 |
| 12 | ENSBTAG00000015563 | PDE12 | 19.27 | 51.96 | 1.43 |
| S.No. | Gene ID | Gene | Cattle Control FPKM Value | Cattle Infected FPKM Value | log2 (Fold Change) |
|---|---|---|---|---|---|
| 1 | ENSBTAG00000003366 | DDX58 | 25.853 | 165.535 | 2.67873 |
| 2 | ENSBTAG00000007554 | IFI6 | 85.1433 | 1240.16 | 3.86449 |
| 3 | ENSBTAG00000007881 | IFIT1 | 63.5862 | 530.603 | 3.06085 |
| 4 | ENSBTAG00000008142 | IFIH1 | 55.5351 | 152.708 | 1.45931 |
| 5 | ENSBTAG00000008471 | MX2 | 34.5429 | 605.862 | 4.13253 |
| 6 | ENSBTAG00000009768 | IFIT3 | 7.60867 | 365.035 | 5.58425 |
| 7 | ENSBTAG00000012529 | IFNG | 11.1059 | 39.2096 | 1.81988 |
| 8 | ENSBTAG00000014707 | ISG15 | 159.192 | 3860.52 | 4.59996 |
| 9 | ENSBTAG00000014762 | ISG20 | 90.9695 | 323.425 | 1.82998 |
| 10 | ENSBTAG00000016061 | RSAD2 | 41.7415 | 623.244 | 3.90024 |
| 11 | ENSBTAG00000017367 | IFIT5 | 22.1536 | 185.959 | 3.06937 |
| 12 | ENSBTAG00000020538 | HERC5 | 41.6011 | 155.056 | 1.89810 |
| 13 | ENSBTAG00000030913 | Mx1 | 50.5075 | 708.813 | 3.81084 |
| 14 | ENSBTAG00000030932 | IFI44L | 49.6875 | 259.9 | 2.38700 |
| 15 | ENSBTAG00000034349 | IFI44 | 35.618 | 295.417 | 3.05208 |
| 16 | ENSBTAG00000034918 | IFIT2 | 14.4535 | 126.911 | 3.13432 |
| 17 | ENSBTAG00000037527 | OAS1X | 56.6369 | 364.978 | 2.68799 |
| 18 | ENSBTAG00000039861 | OAS1Y | 42.7186 | 970.258 | 4.50543 |
| 19 | ENSBTAG00000046580 | DHX58 | 11.4804 | 144.685 | 3.65567 |
| 20 | ENSBTAG00000047680 | IRF7 | 98.2855 | 457.14 | 2.21759 |
| 21 | ENSBTAG00000008703 | EIF2AK2 | 24.6231 | 92.6854 | 1.91233 |
| 22 | ENSBTAG00000019015 | IFITM3 | 215.423 | 683.86 | 1.66653 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tirumurugaan, K.G.; Pawar, R.M.; Dhinakar Raj, G.; Thangavelu, A.; Hammond, J.A.; Parida, S. RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle. Viruses 2020, 12, 463. https://doi.org/10.3390/v12040463
Tirumurugaan KG, Pawar RM, Dhinakar Raj G, Thangavelu A, Hammond JA, Parida S. RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle. Viruses. 2020; 12(4):463. https://doi.org/10.3390/v12040463
Chicago/Turabian StyleTirumurugaan, Krishnaswamy Gopalan, Rahul Mohanchandra Pawar, Gopal Dhinakar Raj, Arthanari Thangavelu, John A. Hammond, and Satya Parida. 2020. "RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle" Viruses 12, no. 4: 463. https://doi.org/10.3390/v12040463
APA StyleTirumurugaan, K. G., Pawar, R. M., Dhinakar Raj, G., Thangavelu, A., Hammond, J. A., & Parida, S. (2020). RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle. Viruses, 12(4), 463. https://doi.org/10.3390/v12040463






