Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors
Abstract
1. Introduction
2. Modules Forming the Siphoviridae Tail Tip
2.1. The TMP-Dit-Tal Triad
2.2. RBPs and Ancillary Proteins
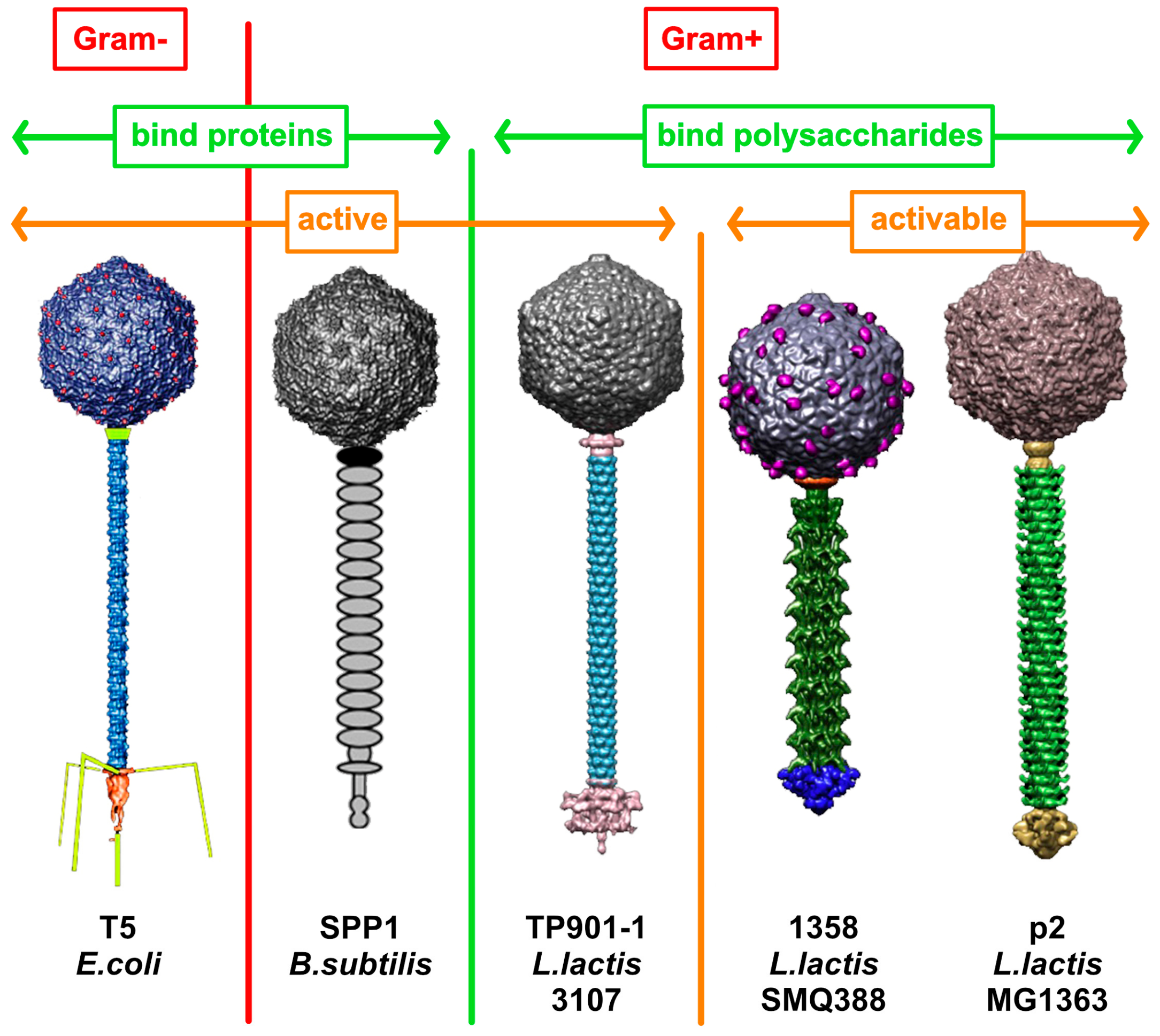
3. Architecture of Siphophage Adhesion Devices Targeting Proteinaceous Receptors
3.1. Gram-Negative Host
3.1.1. Phage lambda
3.1.2. Phage T5
3.2. Gram-Positive Host
3.2.1. Phage SPP1
3.2.2. Lactococcal Phages c2 and bIL67
4. Architecture of Siphophage Adhesion Devices Targeting Hosts’ Surface Polysaccharides
4.1. Binding-Ready Baseplate
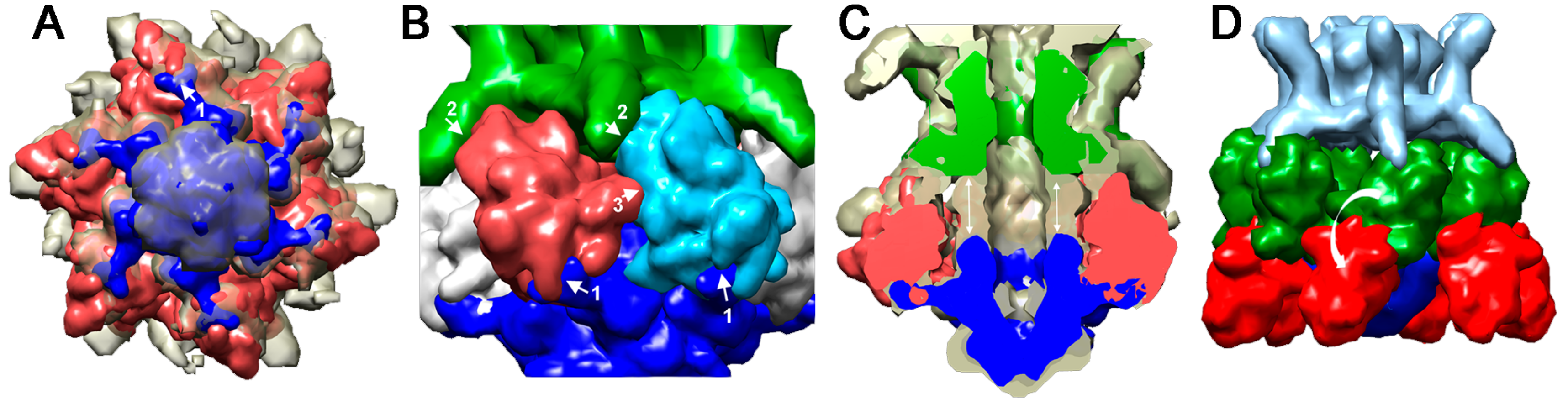
4.1.1. Lactococcal Phage TP901-1
4.1.2. Phage Tuc2009
4.1.3. Listerial Phages A118 and PSA
4.1.4. Staphylococcal Phage 80α
4.2. Activatable Baseplates
4.2.1. Lactococcus lactis Phage p2
4.2.2. Lactococcus lactis Phage 1358
4.2.3. Other Phages
5. Perspectives
Conflicts of Interest
References
- Guenther, S.; Herzig, O.; Fieseler, L.; Klumpp, J.; Loessner, M.J. Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int. J. Food Microbiol. 2012, 154, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Cambillau, C.; van Sinderen, D. Host recognition by lactic acid bacterial phages. FEMS Microbiol. Rev. 2017, 41, S16–S26. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, S.; Leiman, P.G.; Kostyuchenko, V.A.; Chipman, P.R.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure of the cell-puncturing device of bacteriophage T4. Nature 2002, 415, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
- Casjens, S.R.; Hendrix, R.W. Bacteriophage lambda: Early pioneer and still relevant. Virology 2015, 479–480, 310–330. [Google Scholar] [CrossRef]
- Cumby, N.; Reimer, K.; Mengin-Lecreulx, D.; Davidson, A.R.; Maxwell, K.L. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol. Microbiol. 2015, 96, 437–447. [Google Scholar] [CrossRef]
- Zivanovic, Y.; Confalonieri, F.; Ponchon, L.; Lurz, R.; Chami, M.; Flayhan, A.; Renouard, M.; Huet, A.; Decottignies, P.; Davidson, A.R.; et al. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J. Virol. 2014, 88, 1162–1174. [Google Scholar] [CrossRef]
- Mahony, J.; van Sinderen, D. Structural aspects of the interaction of dairy phages with their host bacteria. Viruses 2012, 4, 1410–1424. [Google Scholar] [CrossRef]
- Spinelli, S.; Veesler, D.; Bebeacua, C.; Cambillau, C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front. Microbiol. 2014, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018, 10, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Pluckthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350e4. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ferreira, R.C.; Hupfeld, M.; Nazarov, S.; Taylor, N.M.; Shneider, M.M.; Obbineni, J.M.; Loessner, M.J.; Ishikawa, T.; Klumpp, J.; Leiman, P.G. Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells. EMBO J. 2019, 38, 3. [Google Scholar] [CrossRef]
- Kizziah, J.L.; Manning, K.A.; Dearborn, A.D.; Dokland, T. Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. PLoS Pathogens 2020. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett 2016, 363, 4. [Google Scholar] [CrossRef]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.P.; van Sinderen, D. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. MBio 2014, 5, e00880-14. [Google Scholar] [CrossRef] [PubMed]
- Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Armalyte, J.; Meyrand, M.; Giaouris, E.; Palussiere, S.; Furlan, S.; Pechoux, C.; Ainsworth, S.; et al. Another Brick in the Wall: A Rhamnan Polysaccharide Trapped inside Peptidoglycan of Lactococcus lactis. MBio 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Alqarni, M.; Stockdale, S.; Spinelli, S.; Feyereisen, M.; Cambillau, C.; Sinderen, D.V. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci. Rep. 2016, 6, 36667. [Google Scholar] [CrossRef]
- Vegge, C.S.; Brondsted, L.; Neve, H.; Mc Grath, S.; van Sinderen, D.; Vogensen, F.K. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 2005, 187, 4187–4197. [Google Scholar] [CrossRef]
- Bebeacua, C.; Bron, P.; Lai, L.; Vegge, C.S.; Brondsted, L.; Spinelli, S.; Campanacci, V.; Veesler, D.; van Heel, M.; Cambillau, C. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J. Biol. Chem. 2010, 285, 39079–39086. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Stockdale, S.R.; Collins, B.; Spinelli, S.; Douillard, F.P.; Cambillau, C.; van Sinderen, D. Lactococcus lactis phage TP901-1 as a model for Siphoviridae virion assembly. Bacteriophage 2016, 6, e1123795. [Google Scholar] [CrossRef][Green Version]
- Millen, A.M.; Romero, D.A. Genetic determinants of lactococcal C2viruses for host infection and their role in phage evolution. J. Gen. Virol. 2016, 97, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Sciara, G.; Bebeacua, C.; Bron, P.; Tremblay, D.; Ortiz-Lombardia, M.; Lichiere, J.; van Heel, M.; Campanacci, V.; Moineau, S.; Cambillau, C. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. USA 2010, 107, 6852–6857. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Bebeacua, C.; Orlov, I.; Tremblay, D.; Klaholz, B.P.; Moineau, S.; Cambillau, C. Cryo-electron microscopy structure of lactococcal siphophage 1358 virion. J. Virol. 2014, 88, 8900–8910. [Google Scholar] [CrossRef] [PubMed]
- Brondsted, L.; Ostergaard, S.; Pedersen, M.; Hammer, K.; Vogensen, F.K. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: Evolution, structure, and genome organization of lactococcal bacteriophages. Virology 2001, 283, 93–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seegers, J.F.; Mc Grath, S.; O’Connell-Motherway, M.; Arendt, E.K.; van de Guchte, M.; Creaven, M.; Fitzgerald, G.F.; van Sinderen, D. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 2004, 329, 40–52. [Google Scholar] [CrossRef]
- Lavelle, K.; Goulet, A.; McDonnell, B.; Spinelli, S.; van Sinderen, D.; Mahony, J.; Cambillau, C. Revisiting the Host Adhesion Determinants of Streptococcus thermophilus Siphophages. Microb. Boitech. 2020, in press. [Google Scholar]
- Veesler, D.; Robin, G.; Lichiere, J.; Auzat, I.; Tavares, P.; Bron, P.; Campanacci, V.; Cambillau, C. Crystal Structure of Bacteriophage SPP1 Distal Tail Protein (gp19.1): A Baseplate Hub Paradigm In Gram-Positive Infecting Phages. J. Biol. Chem. 2010, 285, 36666–36673. [Google Scholar] [CrossRef]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichiere, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D.; et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef]
- Flayhan, A.; Vellieux, F.M.; Lurz, R.; Maury, O.; Contreras-Martel, C.; Girard, E.; Boulanger, P.; Breyton, C. Crystal Structure of pb9, the Distal Tail Protein of Bacteriophage T5: A Conserved Structural Motif among All Siphophages. J. Virol. 2014, 88, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Pell, L.G.; Kanelis, V.; Donaldson, L.W.; Howell, P.L.; Davidson, A.R. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA 2009, 106, 4160–4165. [Google Scholar] [CrossRef] [PubMed]
- Jobichen, C.; Chakraborty, S.; Li, M.; Zheng, J.; Joseph, L.; Mok, Y.K.; Leung, K.Y.; Sivaraman, J. Structural basis for the secretion of EvpC: A key type VI secretion system protein from Edwardsiella tarda. PLoS ONE 2010, 5, e12910. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. MMBR 2011, 75, 423–433, first page of table of contents. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordonez, C.L.; Lory, S.; et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Wang, J.; Hofnung, M.; Charbit, A. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 2000, 182, 508–512. [Google Scholar] [CrossRef]
- Vinga, I.; Baptista, C.; Auzat, I.; Petipas, I.; Lurz, R.; Tavares, P.; Santos, M.A.; Sao-Jose, C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 2012, 83, 289–303. [Google Scholar] [CrossRef]
- Stockdale, S.R.; Mahony, J.; Courtin, P.; Chapot-Chartier, M.P.; van Pijkeren, J.P.; Britton, R.A.; Neve, H.; Heller, K.J.; Aideh, B.; Vogensen, F.K.; et al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 2013, 288, 5581–5590. [Google Scholar] [CrossRef]
- Duplessis, M.; Moineau, S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 2001, 41, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.; Ostergaard, S.; Bresciani, J.; Vogensen, F.K. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 2000, 276, 315–328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boulanger, P.; Jacquot, P.; Plancon, L.; Chami, M.; Engel, A.; Parquet, C.; Herbeuval, C.; Letellier, L. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 2008, 283, 13556–13564. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, M.E.; Fina Martin, J.; Duran, R.; Nemirovsky, S.I.; Sanchez Rivas, C.; Bowman, C.; Russell, D.; Hatfull, G.F.; Cambillau, C.; Piuri, M. Characterization of prophages containing “evolved” Dit/Tal modules in the genome of Lactobacillus casei BL23. Appl. Microbiol. Biotechnol. 2016, 100, 9201–9215. [Google Scholar] [CrossRef]
- Dieterle, M.E.; Spinelli, S.; Sadovskaya, I.; Piuri, M.; Cambillau, C. Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: A novel type of anti-receptor widespread among lactic acid bacteria phages. Mol. Microbiol. 2017, 104, 608–620. [Google Scholar] [CrossRef]
- Hayes, S.; Vincentelli, R.; Mahony, J.; Nauta, A.; Ramond, L.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Cambillau, C. Functional carbohydrate binding modules identified in evolved dits from siphophages infecting various Gram-positive bacteria. Mol. Microbiol. 2018, 110, 777–795. [Google Scholar] [CrossRef]
- Hayes, S.; Mahony, J.; Vincentelli, R.; Ramond, L.; Nauta, A.; van Sinderen, D.; Cambillau, C. Ubiquitous Carbohydrate Binding Modules Decorate 936 Lactococcal Siphophage Virions. Viruses 2019, 11, 631. [Google Scholar] [CrossRef]
- Philippe, C.; Levesque, S.; Dion, M.; Tremblay, D.M.; Horvath, P.; Cambillau, C.; Franz, C.; Neve, H.; Fremaux, C.; Heller, K.J.; et al. Genomic characterization of a novel genus of phages infecting Streptococcus thermophilus. Appl. Environ. Microbiol. 2020, in press. [Google Scholar]
- Pell, L.G.; Gasmi-Seabrook, G.M.; Morais, M.; Neudecker, P.; Kanelis, V.; Bona, D.; Donaldson, L.W.; Edwards, A.M.; Howell, P.L.; Davidson, A.R.; et al. The solution structure of the C-terminal Ig-like domain of the bacteriophage lambda tail tube protein. J. Mol. Biol. 2010, 403, 468–479. [Google Scholar] [CrossRef]
- Auzat, I.; Droge, A.; Weise, F.; Lurz, R.; Tavares, P. Origin and function of the two major tail proteins of bacteriophage SPP1. Mol. Microbiol. 2008, 70, 557–569. [Google Scholar] [CrossRef]
- Spinelli, S.; Desmyter, A.; Verrips, C.T.; de Haard, H.J.; Moineau, S.; Cambillau, C. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat. Struct. Mol. Biol. 2006, 13, 85–89. [Google Scholar] [CrossRef]
- Ricagno, S.; Campanacci, V.; Blangy, S.; Spinelli, S.; Tremblay, D.; Moineau, S.; Tegoni, M.; Cambillau, C. Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170. J. Virol. 2006, 80, 9331–9335. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Campanacci, V.; Blangy, S.; Moineau, S.; Tegoni, M.; Cambillau, C. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J. Biol. Chem. 2006, 281, 14256–14262. [Google Scholar] [CrossRef] [PubMed]
- Legrand, P.; Collins, B.; Blangy, S.; Murphy, J.; Spinelli, S.; Gutierrez, C.; Richet, N.; Kellenberger, C.; Desmyter, A.; Mahony, J.; et al. The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules. MBio 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Farenc, C.; Spinelli, S.; Vinogradov, E.; Tremblay, D.; Blangy, S.; Sadovskaya, I.; Moineau, S.; Cambillau, C. Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein. J. Virol. 2014, 88, 7005–7015. [Google Scholar] [CrossRef] [PubMed]
- McCabe, O.; Spinelli, S.; Farenc, C.; Labbe, M.; Tremblay, D.; Blangy, S.; Oscarson, S.; Moineau, S.; Cambillau, C. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 2015, 96, 875–886. [Google Scholar] [CrossRef]
- Koc, C.; Xia, G.; Kuhner, P.; Spinelli, S.; Roussel, A.; Cambillau, C.; Stehle, T. Structure of the host-recognition device of Staphylococcus aureus phage varphi11. Sci Rep. 2016, 6, 27581. [Google Scholar] [CrossRef]
- Chappell, J.D.; Prota, A.E.; Dermody, T.S.; Stehle, T. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002, 21, 1–11. [Google Scholar] [CrossRef]
- Burmeister, W.P.; Guilligay, D.; Cusack, S.; Wadell, G.; Arnberg, N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 2004, 78, 7727–7736. [Google Scholar] [CrossRef]
- Van Raaij, M.J.; Mitraki, A.; Lavigne, G.; Cusack, S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 1999, 401, 935–938. [Google Scholar] [CrossRef]
- Van Raaij, M.J.; Schoehn, G.; Burda, M.R.; Miller, S. Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 2001, 314, 1137–1146. [Google Scholar] [CrossRef]
- Cambillau, C. Bacteriophage module reshuffling results in adaptive host range as exemplified by the baseplate model of listerial phage A118. Virology 2015, 484, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.S.; Logger, L.; Spinelli, S.; Legrand, P.; Huyen Pham, T.T.; Nhung Trinh, T.T.; Cherrak, Y.; Zoued, A.; Desmyter, A.; Durand, E.; et al. Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat. Microbiol. 2017, 2, 17103. [Google Scholar] [CrossRef]
- Park, Y.J.; Lacourse, K.D.; Cambillau, C.; DiMaio, F.; Mougous, J.D.; Veesler, D. Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy. Nat. Commun. 2018, 9, 5385. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.; Pell, L.G.; Bona, D.; Tsai, A.; Dai, X.X.; Edwards, A.M.; Hendrix, R.W.; Maxwell, K.L.; Davidson, A.R. Tail Tip Proteins Related to Bacteriophage lambda gpL Coordinate an Iron-Sulfur Cluster. J. Mol. Biol. 2013, 425, 2450–2462. [Google Scholar] [CrossRef]
- Browning, C.; Shneider, M.M.; Bowman, V.D.; Schwarzer, D.; Leiman, P.G. Phage pierces the host cell membrane with the iron-loaded spike. Structure 2012, 20, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Duda, R.L. Bacteriophage lambda PaPa: Not the mother of all lambda phages. Science 1992, 258, 1145–1148. [Google Scholar] [CrossRef]
- Guan, J.; Ibarra, D.; Zeng, L. The role of side tail fibers during the infection cycle of phage lambda. Virology 2019, 527, 57–63. [Google Scholar] [CrossRef]
- Bartual, S.G.; Otero, J.M.; Garcia-Doval, C.; Llamas-Saiz, A.L.; Kahn, R.; Fox, G.C.; van Raaij, M.J. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl. Acad. Sci. USA 2010, 107, 20287–20292. [Google Scholar] [CrossRef]
- Montag, D.; Schwarz, H.; Henning, U. A component of the side tail fiber of Escherichia coli bacteriophage lambda can functionally replace the receptor-recognizing part of a long tail fiber protein of the unrelated bacteriophage T4. J. Bacteriol. 1989, 171, 4378–4384. [Google Scholar] [CrossRef] [PubMed]
- North, O.I.; Sakai, K.; Yamashita, E.; Nakagawa, A.; Iwazaki, T.; Buttner, C.R.; Takeda, S.; Davidson, A.R. Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres. Nat. Microbiol. 2019, 4, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Doval, C.; Caston, J.R.; Luque, D.; Granell, M.; Otero, J.M.; Llamas-Saiz, A.L.; Renouard, M.; Boulanger, P.; van Raaij, M.J. Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone. Viruses 2015, 7, 6424–6440. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Blangy, S.; Spinelli, S.; Tavares, P.; Campanacci, V.; Cambillau, C. Crystal structure of Bacillus subtilis SPP1 phage gp22 shares fold similarity with a domain of Lactococcal phage p2 RBP. Protein Sci. 2010, 19, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Blangy, S.; Lichiere, J.; Ortiz-Lombarda, M.; Tavares, P.; Campanacci, V.; Cambillau, C. Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. Protein Sci. 2010, 19, 1812–1816. [Google Scholar] [CrossRef]
- Nikolaienko, R.M.; Hammel, M.; Dubreuil, V.; Zalmai, R.; Hall, D.R.; Mehzabeen, N.; Karuppan, S.J.; Harroch, S.; Stella, S.L.; Bouyain, S. Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues. J. Biol. Chem. 2016, 291, 21335–21349. [Google Scholar] [CrossRef]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012, 726, 115–142. [Google Scholar]
- Breyton, C.; Flayhan, A.; Gabel, F.; Lethier, M.; Durand, G.; Boulanger, P.; Chami, M.; Ebel, C. Assessing the conformational changes of pb5, the receptor-binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA. J. Biol. Chem. 2013, 288, 30763–30772. [Google Scholar] [CrossRef]
- Chapot-Chartier, M.P.; Vinogradov, E.; Sadovskaya, I.; Andre, G.; Mistou, M.Y.; Trieu-Cuot, P.; Furlan, S.; Bidnenko, E.; Courtin, P.; Pechoux, C.; et al. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 2010, 285, 10464–10471. [Google Scholar] [CrossRef]
- Baptista, C.; Santos, M.A.; Sao-Jose, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 2008, 190, 4989–4996. [Google Scholar] [CrossRef]
- Godinho, L.M.; El Sadek Fadel, M.; Monniot, C.; Jakutyte, L.; Auzat, I.; Labarde, A.; Djacem, K.; Oliveira, L.; Carballido-Lopez, R.; Ayora, S.; et al. The Revisited Genome of Bacillus subtilis Bacteriophage SPP1. Viruses 2018, 10, 705. [Google Scholar] [CrossRef]
- Goulet, A.; Lai-Kee-Him, J.; Veesler, D.; Auzat, I.; Robin, G.; Shepherd, D.A.; Ashcroft, A.E.; Richard, E.; Lichiere, J.; Tavares, P.; et al. The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J. Biol. Chem. 2011, 286, 25397–25405. [Google Scholar] [CrossRef]
- Plisson, C.; White, H.E.; Auzat, I.; Zafarani, A.; Sao-Jose, C.; Lhuillier, S.; Tavares, P.; Orlova, E.V. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007, 26, 3720–3728. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nature Protocols 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Labrie, S.J.; Chopin, M.C.; Moineau, S. Biodiversity and classification of lactococcal phages. Appl Environ. Microbiol. 2006, 72, 4338–4346. [Google Scholar] [CrossRef] [PubMed]
- Lubbers, M.W.; Waterfield, N.R.; Beresford, T.P.; Le Page, R.W.; Jarvis, A.W. Sequencing and analysis of the prolate-headed Lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 1995, 61, 4348–4356. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Veesler, D.; Lichiere, J.; Blangy, S.; Sciara, G.; Moineau, S.; van Sinderen, D.; Bron, P.; Cambillau, C. Solution and electron microscopy characterization of Lactococcal phage baseplates expressed in Escherichia coli. J. Struct. Biol. 2010, 172, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, D.A.; Veesler, D.; Lichiere, J.; Ashcroft, A.E.; Cambillau, C. Unraveling Lactococcal phage baseplate assembly by mass spectrometry. Mol. Cell Proteomics 2011, 10, M111-009787. [Google Scholar] [CrossRef]
- Sciara, G.; Blangy, S.; Siponen, M.; Mc Grath, S.; van Sinderen, D.; Tegoni, M.; Cambillau, C.; Campanacci, V. A topological model of the baseplate of lactococcal phage Tuc2009. J. Biol. Chem. 2008, 283, 2716–2723. [Google Scholar] [CrossRef]
- Collins, B.; Bebeacua, C.; Mahony, J.; Blangy, S.; Douillard, F.P.; Veesler, D.; Cambillau, C.; van Sinderen, D. Structure and functional analysis of the host recognition device of Lactococcal phage tuc2009. J. Virol. 2013, 87, 8429–8440. [Google Scholar] [CrossRef]
- Hayes, S.; Duhoo, Y.; Neve, H.; Murphy, J.; Noben, J.P.; Franz, C.; Cambillau, C.; Mahony, J.; Nauta, A.; van Sinderen, D. Identification of Dual Receptor Binding Protein Systems in Lactococcal 936 Group Phages. Viruses 2018, 10, 668. [Google Scholar] [CrossRef] [PubMed]
- Bebeacua, C.; Tremblay, D.; Farenc, C.; Chapot-Chartier, M.P.; Sadovskaya, I.; van Heel, M.; Veesler, D.; Moineau, S.; Cambillau, C. Structure, adsorption to host, and infection mechanism of virulent Lactococcal phage p2. J. Virol. 2013, 87, 12302–12312. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, M.E.; Moineau, S. Genome organization and characterization of the virulent Lactococcal phage 1358 and its similarities to Listeria phages. Appl Environ. Microbiol. 2010, 76, 1623–1632. [Google Scholar] [CrossRef]
- Mahony, J.; Oliveira, J.; Collins, B.; Hanemaaijer, L.; Lugli, G.A.; Neve, H.; Ventura, M.; Kouwen, T.R.; Cambillau, C.; van Sinderen, D. Genetic and functional characterisation of the Lactococcal P335 phage-host interactions. BMC Genomics 2017, 18, 146. [Google Scholar] [CrossRef] [PubMed]





| Query | Target (PDB ID, Source) | Probability (%) | E-Value | Identity (%) | Query Residue Range (total) | Target Residue Range (Total) |
|---|---|---|---|---|---|---|
| lambda gpM (Dit) | 6F2M, T5 | 94 | 0.42 | 11 | 4–77 (109) | 12–85 (217) |
| lambda gpJ (Tal) | 3D37, N. meningitidis MC58 | 95.9 | 0.18 | 14 | 327–499 (1132) | 143–320 (381) |
| 3CDD, Shewanella MuSo2 | 95.8 | 0.18 | 10 | 326–499 | 146–324 | |
| 1WRU, phage Mu | 94.1 | 0.69 | 16 | 326–499 | 140–318 | |
| 3GS9, L. monocytogenes A118 | 85.0 | 12 | 8 | 334–499 | 149–307 | |
| 5UTK, Receptor extracellular region (fibronectin III repeats) | 98.7 | 1.6e-6 | 11 | 570–811 | 64–297 | |
| T5 pb3 (Tal) | 3D37, N. meningitidis MC58 | 97.3 | 0.012 | 11 | 529–725 (949) | 143–350 (381) |
| 3CDD, Shewanella MuSo2 | 97.1 | 0.011 | 12 | 528–709 | 146–339 (361) | |
| 1WRU, phage Mu | 96.8 | 0.026 | 15 | 529–716 | 141–339 (379) | |
| 3GS9, Listeria | 91.3 | 4.3 | 15 | 533–708 | 146–320 (342) | |
| 5E53, adhesion molecule (fibronectin III repeats) | 98.6 | 1.9e-5 | 13 | 690–949 | 133–295 | |
| SPP1 gp21 (Tal) | 3GS9, L. monocytogenes A118 | 99.9 | 4.9e-25 | 12 | 3–404 (1111) | 1–141 (342) |
| 3CDD, Shewanella MuSo2 | 98.9 | 4.1e-7 | 11 | 1–382 | 2–340 | |
| 3D37, N. meningitidis MC58 | 98.9 | 7.7e-7 | 10 | 1–381 | 2–336 | |
| 1WRU, phage Mu | 98.9 | 9.2e-7 | 11 | 2–381 | 1–334 | |
| 5E53, adhesion molecule (fibronectin III repeats) | 96.4 | 0.011 | 14 | 422–518 | 204–301 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goulet, A.; Spinelli, S.; Mahony, J.; Cambillau, C. Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors. Viruses 2020, 12, 512. https://doi.org/10.3390/v12050512
Goulet A, Spinelli S, Mahony J, Cambillau C. Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors. Viruses. 2020; 12(5):512. https://doi.org/10.3390/v12050512
Chicago/Turabian StyleGoulet, Adeline, Silvia Spinelli, Jennifer Mahony, and Christian Cambillau. 2020. "Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors" Viruses 12, no. 5: 512. https://doi.org/10.3390/v12050512
APA StyleGoulet, A., Spinelli, S., Mahony, J., & Cambillau, C. (2020). Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors. Viruses, 12(5), 512. https://doi.org/10.3390/v12050512






