SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions
Abstract
1. Introduction
2. Epidemiology and Transmission of SARS-CoV-2
- SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2
- COVID-19 or Covid-19: Corona virus disease, 2019. COVID-19 is the official name of the disease manifested by SARS-CoV-2.
- R0: Reproduction number that defines the number of secondary cases that will be produced by a single infectious index case in a population that is fully susceptible to the disease. For example, a R0 of 2 means that, on average, one primary index case would infect two other people, generating two secondary cases. Continuous horizontal (human-to-human) transmission will occur if R0 is above the critical threshold of one.
- Fomite Transmission: A fomite is any inanimate object (i.e., surface) when contaminated with or exposed to infectious agent, can serve as a source to transmit the agent into a new host.
- Non-Pharmacological Interventions (NPIs): NPIs are evidence based, non-invasive, mostly policy/regulation driven interventions on human health. NPIs (i.e., physical [“social”] distancing) can be very effective to contain viral shedding.
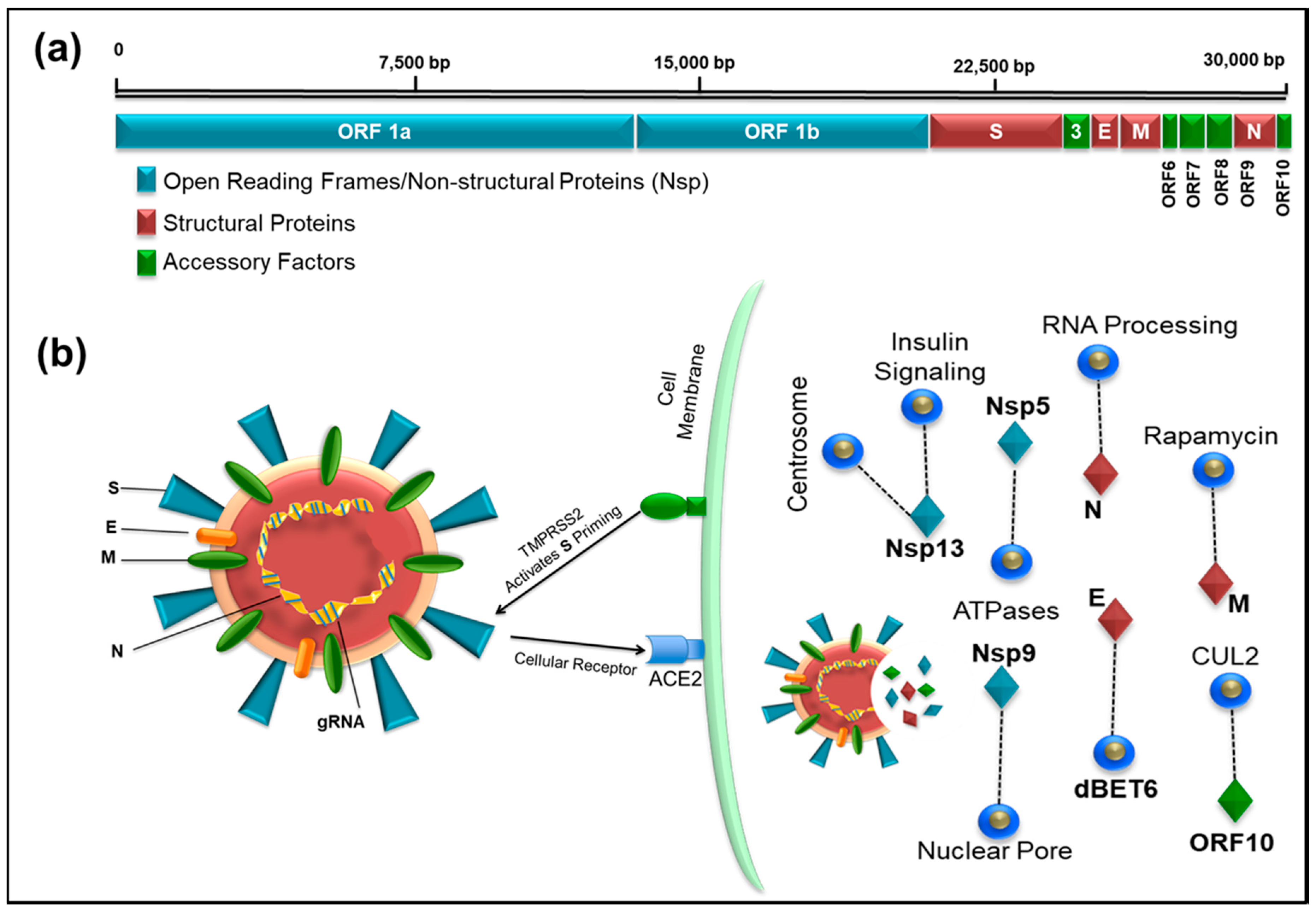
3. Genomics of SARS-CoV-2
4. Transcriptomic Map and SARS CoV-2-Human Protein–Protein Interactions to Identify Drug Targets
5. Diagnosis of COVID-19
6. Development of Vaccines and Experimental Therapeutic Interventions for SARS-CoV-2
6.1. Vaccine Development
6.2. Experimental Therapeutic Interventions
6.2.1. Convalescent Plasma (CP) Therapy
6.2.2. Soluble Human Angiotensin-Converting Enzyme 2 (ACE2)
7. Drug Repurposing for COVID-19
- Lopinavir (LPV)-Ritonavir (RTV) combination (Kaletra): This is an FDA-approved drug for HIV-1 treatment. Lopinavir is a protease inhibitor that inhibits virus particle maturation, a late step in HIV-1 replication, while ritonavir helps boost the activity of lopinavir by inhibiting CYP3A enzymes that slows down the rate at which lopinavir is broken down in the liver [69]. Findings from in vitro and animal studies against both SARS and MERS indicate its potential for COVID-19 treatment [69,70,71,72]. Lopinavir-Ritonavir has been used either on its own or in combination with either alpha interferon (China) or chloroquine/hydroxychloroquine (South Korea) for COVID-19 treatment with some success [73,74]. However, new data from China cast doubt on the beneficial effect in seriously ill COVID-19 patients [75]. Thus, results from additional clinical trials are needed to establish the efficacy of this treatment for COVID-19 which are currently underway.
- Favipiravir (Favilavir or Avigan): Favipiravir (FPV) is an RNA-dependent RNA polymerase inhibitor developed by Fujifilm Toyama Chemical in Japan that is safe and has been effective in other viral infections, including influenza [76,77]. It has now been shown to be useful against SARS-CoV-2 in initial clinical trials conducted in Wuhan and Shenzhen [78]. In this study, the effects of FPV versus LPV/RTV were compared during the treatment of COVID-19 patients. The FPV-treated patients demonstrated much better therapeutic response especially with regard to faster viral clearance and improvement rate in chest imaging. Based on these encouraging results, favipiravir has been approved by the National Medical Products Administration of China as the first anti-COVID-19 drug in the country [66].
- Chloroquine/Hydroxychloroquine: Chloroquine is an inexpensive drug for the treatment of malaria and features on the WHO list of essential medicines. It is also used as an anti-inflammatory agent for the treatment of autoimmune diseases. Chloroquine is thought to inhibit virus replication by increasing endosomal pH as many viruses such as Ebola and Marburg that require the acidic environment of the endosome for successful replication [79,80,81]. However, a recent study showed that the anti-inflammatory effects of chloroquine are mediated by upregulation of the cyclin-dependent kinase inhibitor, p21 [82]. In vitro studies have shown its potent antiviral effect against the SARS-CoV-2 [83]. A multicenter clinical trial in China has reported efficacy with amelioration of exacerbation of pneumonia and acceptable safety margin with use of chloroquine for treatment of COVID-19 [10]. Hydroxychloroquine is an analogue of chloroquine which is more stable with better clinical safety profile and has anti-SARS-CoV-2 activity. It has been shown to quicken recovery and clearance of the virus in COVID-19 patients and used successfully in combination with the macrolide antibiotic azithromycin [84]. A recent clinical trial, however, has shown disappointing results with the combination of azithromycin with hydroxychloroquine in critically-ill COVID-19 patients [85], suggesting that larger studies with controlled design are needed before conclusive recommendations can be made for chloroquine/hydroxychloroquine in the treatment of COVID-19. Interestingly, chloroquine and hydroxychloroquine are zinc ionophores and zinc has been shown to inhibit RNA-dependent RNA polymerase enzyme of coronaviruses [86,87]. Thus, one reason for the limited success of some of these clinical trials could be due to absence of zinc supplementation which may be necessary to observe the therapeutic effects of these drugs on SARS-CoV-2 and other RNA virus infections [88].
- Remdesivir (GS-5734): Remdesivir is a nucleotide analogue prodrug with broad spectrum antiviral activity against many RNA viruses [89]. Like Favipiravir, it blocks RNA-dependent RNA polymerase, an enzyme that replicates the viral genome, inhibiting an early step in virus replication, compared to protease inhibitors that target the late steps of virus replication [90,91]. It has also shown to inhibit replication of MERSCoV, SARS-CoV, and SARS-CoV-2 in animal models [83,89,92,93]. So far, it has been used as an investigational drug for the treatment of Ebola, MERS-CoV, and SARS-CoV2, and other RNA viruses, but has not been approved for any disease [83,89,92,93]. In a compassionate use of remdesivir in a cohort of patients hospitalized for severe COVID-19, the developers of the drug (Gilead Sciences, City, US State abbrev., USA) reported clinical improvement in 68% (36 of 53) of patients [94]. The first randomized, double-blind, placebo-controlled, multicenter clinical trial of remdesivir in 237 patients from Hubei, China, has just been published [95]. Unfortunately, it did not show statistically meaningful clinical benefits except for numerical reduction in time to clinical improvement [95]. Furthermore, treatment with remdesivir had to be stopped early in some patients because of undesirable effects in 12% patients versus 5% patients on placebo. Similar results have been announced from the first US clinical trial of the drug at the time of this writing, which are still unpublished. Further results are awaited on multiple clinical trials of remdesivir in several countries for more conclusive guidelines on its use in COVID-19 patients.
- SNG001: SNG001 is an inhaled experimental drug (interferon beta) developed by the UK biotech firm Synairgen. The ability to inhale the drug will allow the patients to “self-administer” it by using a small hand-held nebulizer. It was developed for the severe lung disease chronic-obstructive pulmonary disorder (COPD), but, due to the current COVID-19 crisis, it has been fast-tracked for use in a 100-patient phase II clinical trial (EudraCT2020-001023-14) in the UK (https://adisinsight.springer.com/drugs/800024480), the results of which are awaited.
- Tocilizumab: Tocilizumab is a humanized monoclonal antibody against the interleukin-6 receptor (IL-6R) that is approved by FDA to treat patients with rheumatoid arthritis, systemic juvenile idiopathic arthritis, and giant cell arteritis [96]. IL-6 has been shown to be a key mediator of cytokine release storm (CRS) observed in critically ill COVID-19 [96]. Therefore, it has been proposed as a potential therapy to treat such patients [97]. Thus, Tocilizumab has recently been used as an immunosuppressive agent during CRS observed in severely ill COVID-19 patients in China and Italy with promising results [98,99]. COVID-19 patients treated with Tocilizumab in China showed marked improvement indicating that Tocilizumab potentially could be very effective in treating patients with severe infection. Consistent with this, administration of tocilizumab in a COVID-19 patient with pneumonia in Italy showed favorable changes of CT findings within 14 days of treatment [100]. It is turning out to be a promising therapy to treat severely ill COVID-19 patients.
- Kinases: p21-activated protein kinases (PAKs) are cytosolic serine/threonine protein kinases downstream of small (p21) GTPases, including members of the Cdc42 and Rac families. Multiple studies have shown that the major pathogenic kinase in this group, PAK1, plays an important role in the entry, replication and spread of several important viruses, including influenza and HIV [101,102]. Coronaviruses exploit macropinocytosis to gain entry into cells and this process has been shown to be dependent on PAK1 activity [103,104]. Targeting of PAK1 to prevent micropinocytosis has been implicated for therapeutic intervention [105]. This strongly suggests that PAK1-inhibitors could be valuable for the treatment of COVID-19 infection. PAK-1 inhibitors include caffeic acid and its ester, propolis, ketorolac, and triptolide. Unfortunately, all these have problems with solubility and cell penetration. However, newer PAK-1 inhibitors, such as 15K (the 1,2,3-triazolyl ester of ketorolac, that is 500 times more potent at inhibiting PAK1 than the parent compound [106], minnelide (in which a hydroxyl group of triptolide is phosphorylated, boosting its water-solubility over 3000 times [107], and frondoside A [108] are much more potent and may be of value in suppressing the effects of this virus.
8. Non-Pharmacological Interventions
9. Future Directions for COVID-19 Research
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization, WHO. WHO Director-General‘s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 9 May 2020).
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- John Hopkins University of Medicine. Coronavirus Resource Center John Hopkins University of Medicine. 2020. Available online: https://coronavirus.jhu.edu/map.html (accessed on 9 May 2020).
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Covid-19 National Emergency Response Center, E.; Case Management Team, K.C.F.D.C. Prevention. Early Epidemiological and Clinical Characteristics of 28 Cases of Coronavirus Disease in South Korea. Osong Public Health Res. Perspect 2020, 11, 8–14. [Google Scholar] [CrossRef]
- Center for Disease Control, U. Severe Outcomes among Patients with Coronavirus Disease Disease 2019 (COVID-19)—United States, February 12-March 16, 2020. Morb. Mortal. Wkly. Rep. (MMWR) 2020, 69, 343–346. [Google Scholar] [CrossRef]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Sanche, S.; Lin, Y.T.; Xu, C.; Romero-Severson, E.; Hengartner, N.; Ke, R. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26. in press. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, Y.Q. On a Statistical Transmission Model in Analysis of the Early Phase of COVID-19 Outbreak. Stat. Biosci. 2020, 1, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Dong, Y.; Yan, H.; Zhao, C.; Li, X.; Liu, W.; He, M.; Tang, S.; Xi, S. Impact of temperature on the dynamics of the COVID-19 outbreak in China. Sci. Total Environ. 2020, 728, 138890. [Google Scholar] [CrossRef] [PubMed]
- Sobral, M.F.F.; Duarte, G.B.; da Penha Sobral, A.I.G.; Marinho, M.L.M.; de Souza Melo, A. Association between climate variables and global transmission oF SARS-CoV-2. Sci. Total Environ. 2020, 729, 138997. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.; Hui, K.P.Y.; Yen, H.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, in press. [Google Scholar] [CrossRef]
- Morawska, L.; Cao, J. Airborne transmission of SARS-CoV-2: The world should face the reality. Environ. Int. 2020, 139, 105730. [Google Scholar] [CrossRef]
- Setti, L.; Passarini, F.; De Gennaro, G.; Barbieri, P.; Perrone, M.G.; Borelli, M.; Palmisani, J.; Di Gilio, A.; Piscitelli, P.; Miani, A. Airborne Transmission Route of COVID-19: Why 2 Meters/6 Feet of Inter-Personal Distance Could Not Be Enough. Int. J. Environ. Res. Public Health 2020, 17, 2932. [Google Scholar] [CrossRef]
- Burke, R.M.; Midgley, C.M.; Dratch, A.; Fenstersheib, M.; Haupt, T.; Holshue, M.; Ghinai, I.; Jarashow, M.C.; Lo, J.; McPherson, T.D.; et al. Active Monitoring of Persons Exposed to Patients with Confirmed COVID-19—United States, January–February 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 245–246. [Google Scholar] [CrossRef] [PubMed]
- Casanova, L.M.; Jeon, S.; Rutala, W.A.; Weber, D.J.; Sobsey, M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl. Environ. Microbiol. 2010, 76, 2712–2717. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.S.; McIntosh, K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005, 24, S223–S227. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ’host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. Nature 2020, in press. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Ceraolo, C.; Giorgi, F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020, 92, 522–528. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Curr. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, in press. [Google Scholar] [CrossRef]
- Liu, P.; Chen, W.; Chen, J.P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef]
- Kautz, T.F.; Forrester, N.L. RNA Virus Fidelity Mutants: A Useful Tool for Evolutionary Biology or a Complex Challenge? Viruses 2018, 10, 600. [Google Scholar] [CrossRef]
- Smith, E.C. The not-so-infinite malleability of RNA viruses: Viral and cellular determinants of RNA virus mutation rates. PLoS Pathog. 2017, 13, e1006254. [Google Scholar] [CrossRef]
- Xiao, Y.; Rouzine, I.M.; Bianco, S.; Acevedo, A.; Goldstein, E.F.; Farkov, M.; Brodsky, L.; Andino, R. RNA Recombination Enhances Adaptability and Is Required for Virus Spread and Virulence. Cell Host Microbe 2016, 19, 493–503. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 1–6. [Google Scholar] [CrossRef]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Stefanelli, P.; Faggioni, G.; Lo Presti, A.; Fiore, S.; Marchi, A.; Benedetti, E.; Fabiani, C.; Anselmo, A.; Ciammaruconi, A.; Fortunato, A.; et al. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: Additional clues on multiple introductions and further circulation in Europe. Eurosurveillance 2020, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 1–10. [Google Scholar] [CrossRef]
- Luan, J.; Lu, Y.; Jin, X.; Zhang, L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020, in press. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Ong, S.W.X.; Tan, Y.K.; Chia, P.Y.; Lee, T.H.; Ng, O.T.; Wong, M.S.Y.; Marimuthu, K. Air, Surface Environmental, and Personal Protective Equipment Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) From a Symptomatic Patient. JAMA 2020, 323, 1610–1612. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X.; Wen, F.; Huang, X.; Ning, G.; Wang, W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Wang, Y.; Kang, H.; Liu, X.; Tong, Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020, 92, 538–539. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, D.; Amanat, F.; Chromikova, V.; Jiang, K.; Strohmeier, S.; Arunkumar, G.A.; Tan, J.; Bhavsar, D.; Capuano, C.; Kirkpatrick, E.; et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57, e100. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. The race for coronavirus vaccines: A graphical guide. Nature 2020, 580, 576–577. [Google Scholar] [CrossRef]
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef]
- Shang, W.; Yang, Y.; Rao, Y.; Rao, X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines 2020, 5, 1–3. [Google Scholar] [CrossRef]
- Hodgson, J. The pandemic pipeline. Nat. Biotechnol. 2020, in press. [Google Scholar] [CrossRef]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gomez Roman, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef]
- Cheng, Y.; Wong, R.; Soo, Y.O.; Wong, W.S.; Lee, C.K.; Ng, M.H.; Chan, P.; Wong, K.C.; Leung, C.B.; Cheng, G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 44–46. [Google Scholar] [CrossRef]
- Hung, I.F.; To, K.K.; Lee, C.K.; Lee, K.L.; Chan, K.; Yan, W.W.; Liu, R.; Watt, C.L.; Chan, W.M.; Lai, K.Y.; et al. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011, 52, 447–456. [Google Scholar] [CrossRef]
- Ko, J.H.; Seok, H.; Cho, S.Y.; Ha, Y.E.; Baek, J.Y.; Kim, S.H.; Kim, Y.J.; Park, J.K.; Chung, C.R.; Kang, E.S.; et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: A single centre experience. Antivir. Ther. 2018, 23, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Wang, Z.; Zhao, F.; Yang, Y.; Li, J.; Yuan, J.; Wang, F.; Li, D.; Yang, M.; Xing, L.; et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA 2020, 323, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, in press. [Google Scholar] [CrossRef]
- de Wilde, A.H.; Jochmans, D.; Posthuma, C.C.; Zevenhoven-Dobbe, J.C.; van Nieuwkoop, S.; Bestebroer, T.M.; van den Hoogen, B.G.; Neyts, J.; Snijder, E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014, 58, 4875–4884. [Google Scholar] [CrossRef]
- Chan, J.F.; Yao, Y.; Yeung, M.L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment With Lopinavir/Ritonavir or Interferon-beta1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef]
- Chan, K.S.; Lai, S.T.; Chu, C.M.; Tsui, E.; Tam, C.Y.; Wong, M.M.; Tse, M.W.; Que, T.L.; Peiris, J.S.; Sung, J.; et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: A multicentre retrospective matched cohort study. Hong Kong Med. J. 2003, 9, 399–406. [Google Scholar]
- Chu, C.M.; Cheng, V.C.; Hung, I.F.; Wong, M.M.; Chan, K.H.; Chan, K.S.; Kao, R.Y.; Poon, L.L.; Wong, C.L.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef]
- Lim, J.; Jeon, S.; Shin, H.Y.; Kim, M.J.; Seong, Y.M.; Lee, W.J.; Choe, K.W.; Kang, Y.M.; Lee, B.; Park, S.J. Case of the Index Patient Who Caused Tertiary Transmission of COVID-19 Infection in Korea: The Application of Lopinavir/Ritonavir for the Treatment of COVID-19 Infected Pneumonia Monitored by Quantitative RT-PCR. J. Korean Med. Sci. 2020, 35, e79. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Liu, X.; Zhu, Y.; Huang, L.; Dan, B.; Zhang, G.; Gao, Y. Factors associated with prolonged viral shedding and impact of Lopinavir/Ritonavir treatment in patients with SARS-CoV-2 infection. MedRxiv 2020. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Smee, D.F.; Tarbet, E.B.; Furuta, Y.; Morrey, J.D.; Barnard, D.L. Synergistic combinations of favipiravir and oseltamivir against wild-type pandemic and oseltamivir-resistant influenza A virus infections in mice. Future Virol. 2013, 8, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020, in press. [Google Scholar] [CrossRef]
- Akpovwa, H. Chloroquine could be used for the treatment of filoviral infections and other viral infections that emerge or emerged from viruses requiring an acidic pH for infectivity. Cell Biochem. Funct. 2016, 34, 191–196. [Google Scholar] [CrossRef]
- Dowall, S.D.; Bosworth, A.; Watson, R.; Bewley, K.; Taylor, I.; Rayner, E.; Hunter, L.; Pearson, G.; Easterbrook, L.; Pitman, J.; et al. Chloroquine inhibited Ebola virus replication in vitro but failed to protect against infection and disease in the in vivo guinea pig model. J. Gen. Virol. 2015, 96, 3484–3492. [Google Scholar] [CrossRef]
- Li, C.; Zhu, X.; Ji, X.; Quanquin, N.; Deng, Y.Q.; Tian, M.; Aliyari, R.; Zuo, X.; Yuan, L.; Afridi, S.K.; et al. Chloroquine, a FDA-approved Drug, Prevents Zika Virus Infection and its Associated Congenital Microcephaly in Mice. EBioMedicine 2017, 24, 189–194. [Google Scholar] [CrossRef]
- Oh, S.; Shin, J.H.; Jang, E.J.; Won, H.Y.; Kim, H.K.; Jeong, M.G.; Kim, K.S.; Hwang, E.S. Anti-inflammatory activity of chloroquine and amodiaquine through p21-mediated suppression of T cell proliferation and Th1 cell differentiation. Biochem. Biophys. Res. Commun. 2016, 474, 345–350. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; Dupont, H.T.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.M.; Delaugerre, C.; Le Goff, J.; Mela-Lima, B.; Ponscarme, D.; Goldwirt, L.; de Castro, N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Subramani, C.; Anang, S.; Muthumohan, R.; Shalimar; Nayak, B.; Ranjith-Kumar, C.T.; Surjit, M. Zinc Salts Block Hepatitis E Virus Replication by Inhibiting the Activity of Viral RNA-Dependent RNA Polymerase. J. Virol. 2017, 91, e00754-17. [Google Scholar] [CrossRef]
- Liang, S.H.; Southon, A.G.; Fraser, B.H.; Krause-Heuer, A.M.; Zhang, B.; Shoup, T.M.; Lewis, R.; Volitakis, I.; Han, Y.; Greguric, I.; et al. Novel Fluorinated 8-Hydroxyquinoline Based Metal Ionophores for Exploring the Metal Hypothesis of Alzheimer’s Disease. ACS Med. Chem. Lett. 2015, 6, 1025–1029. [Google Scholar] [CrossRef]
- Shittu, M.O.; Afolami, O.I. Improving the efficacy of Chloroquine and Hydroxychloroquine against SARS-CoV-2 may require Zinc additives—A better synergy for future COVID-19 clinical trials. Infez. Med. 2020, 28, 192–197. [Google Scholar]
- Sheahan, T.P.; Sims, A.C.; Graham, R.L.; Menachery, V.D.; Gralinski, L.E.; Case, J.B.; Leist, S.R.; Pyrc, K.; Feng, J.Y.; Trantcheva, I.; et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017, 9, 396. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Gotte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, in press. [Google Scholar] [CrossRef]
- Lo, M.K.; Jordan, R.; Arvey, A.; Sudhamsu, J.; Shrivastava-Ranjan, P.; Hotard, A.L.; Flint, M.; McMullan, L.K.; Siegel, D.; Clarke, M.O.; et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7, 43395. [Google Scholar] [CrossRef]
- de Wit, E.; Feldmann, F.; Cronin, J.; Jordan, R.; Okumura, A.; Thomas, T.; Scott, D.; Cihlar, T.; Feldmann, H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2020, 117, 6771–6776. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schafer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, in press. [Google Scholar] [CrossRef]
- Zhang, S.; Li, L.; Shen, A.; Chen, Y.; Qi, Z. Rational Use of Tocilizumab in the Treatment of Novel Coronavirus Pneumonia. Clin. Drug Investig. 2020, in press. [Google Scholar] [CrossRef]
- Ortiz-Martinez, Y. Tocilizumab: A new opportunity in the possible therapeutic arsenal against COVID-19. Travel Med. Infect. Dis. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, Z.; Li, J.W.; Zhao, H.; Wang, G.Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents 2020, in press. [Google Scholar] [CrossRef]
- Zhao, M. Cytokine storm and immunomodulatory therapy in COVID-19: Role of chloroquine and anti-IL-6 monoclonal antibodies. Int. J. Antimicrob. Agents 2020, in press. [Google Scholar] [CrossRef]
- Cellina, M.; Orsi, M.; Bombaci, F.; Sala, M.; Marino, P.; Oliva, G. Favorable changes of CT findings in a patient with COVID-19 pneumonia after treatment with tocilizumab. Diagn. Interv. Imag. 2020, in press. [Google Scholar] [CrossRef]
- Pascua, P.N.; Lee, J.H.; Song, M.S.; Park, S.J.; Baek, Y.H.; Ann, B.H.; Shin, E.Y.; Kim, E.G.; Choi, Y.K. Role of the p21-activated kinases (PAKs) in influenza A virus replication. Biochem. Biophys. Res. Commun. 2011, 414, 569–574. [Google Scholar] [CrossRef]
- Van den Broeke, C.; Radu, M.; Chernoff, J.; Favoreel, H.W. An emerging role for p21-activated kinases (Paks) in viral infections. Trends Cell Biol. 2010, 20, 160–169. [Google Scholar] [CrossRef]
- Burkard, C.; Verheije, M.H.; Wicht, O.; van Kasteren, S.I.; van Kuppeveld, F.J.; Haagmans, B.L.; Pelkmans, L.; Rottier, P.J.; Bosch, B.J.; de Haan, C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014, 10, e1004502. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.C.; Peek, C.T.; Becker, M.M.; Smith, E.C.; Denison, M.R. Coronaviruses induce entry-independent, continuous macropinocytosis. mBio 2014, 5, e01340-14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Simmons, G. Development of novel entry inhibitors targeting emerging viruses. Expert Rev. Anti Infect. Ther. 2012, 10, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.C.Q.; Takahashi, H.; Uto, Y.; Shahinozzaman, M.D.; Tawata, S.; Maruta, H. 1,2,3-Triazolyl ester of Ketorolac: A “Click Chemistry”-based highly potent PAK1-blocking cancer-killer. Eur. J. Med. Chem. 2017, 126, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Lis, L.G.; Schumacher, R.J.; Norris, B.J.; Morgan, M.L.; Cuellar, R.A.; Blazar, B.R.; Suryanarayanan, R.; Gurvich, V.J.; Georg, G.I. Phosphonooxymethyl Prodrug of Triptolide: Synthesis, Physicochemical Characterization, and Efficacy in Human Colon Adenocarcinoma and Ovarian Cancer Xenografts. J. Med. Chem. 2015, 58, 9334–9344. [Google Scholar] [CrossRef]
- Nguyen, B.C.Q.; Yoshimura, K.; Kumazawa, S.; Tawata, S.; Maruta, H. Frondoside A from sea cucumber and nymphaeols from Okinawa propolis: Natural anti-cancer agents that selectively inhibit PAK1 in vitro. Drug Discov. Ther. 2017, 11, 110–114. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Laydon, D.; Nedjati-Gilani, G.; Natsuko Imai, K.A.; Baguelin, M.; Bhatia, S.; Boonyasiri, A.; Cucunubá, Z.; Cuomo-Dannenburg, G.; Dighe, A.; et al. Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID-19 Mortality and Healthcare Demand. Available online: https://www.imperial.ac.uk/media/imperial-college/medicine/sph/ide/gida-fellowships/Imperial-College-COVID19-NPI-modelling-16-03-2020.pdf (accessed on 9 May 2020).
- Gursel, M.; Gursel, I. Is Global BCG Vaccination Coverage Relevant To The Progression Of SARS-CoV-2 Pandemic? Med. Hypotheses 2020, in press. [Google Scholar] [CrossRef]
- Ozdemir, C.; Kucuksezer, U.C.; Tamay, Z.U. Is BCG vaccination effecting the spread and severity of COVID-19? Allergy 2020, in press. [Google Scholar] [CrossRef]
- Long, J.B.; Ehrenfeld, J.M. The Role of Augmented Intelligence (AI) in Detecting and Preventing the Spread of Novel Coronavirus. J. Med. Syst. 2020, 44, 59. [Google Scholar] [CrossRef]
| Category | Data Type | Database |
|---|---|---|
| SARS-CoV-2 Genome Sequencing Data | DNA Sequencing Data | https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ |
| SARS-CoV-2 Transcriptomic Map | RNA Sequencing Data | Open Science Framework: accession number doi:10.17605/OSF.IO/8F6N9 |
| SARS-CoV-2 and Human Protein Interactions | Mass Spectrometry Raw Data | http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD018117 |
| SARS-CoV-2 Strains | Genomic Epidemiology | https://nextstrain.org/ncov https://www.gisaid.org/ |
| The COVID-19 Host Genetics Initiative | Host Genetics Data (GWAS, WES, WGS) | https://www.covid19hg.org/ |
| COVID-19 Cell Atlas | Single cell transcriptomics data | www.covid19cellatlas.org |
| List of Clinical Trials | Clinical Trial Related Information | https://clinicaltrials.gov/ct2/home |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, M.; Mustafa, F.; Rizvi, T.A.; Loney, T.; Al Suwaidi, H.; Al-Marzouqi, A.H.H.; Kamal Eldin, A.; Alsabeeha, N.; Adrian, T.E.; Stefanini, C.; et al. SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses 2020, 12, 526. https://doi.org/10.3390/v12050526
Uddin M, Mustafa F, Rizvi TA, Loney T, Al Suwaidi H, Al-Marzouqi AHH, Kamal Eldin A, Alsabeeha N, Adrian TE, Stefanini C, et al. SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses. 2020; 12(5):526. https://doi.org/10.3390/v12050526
Chicago/Turabian StyleUddin, Mohammed, Farah Mustafa, Tahir A. Rizvi, Tom Loney, Hanan Al Suwaidi, Ahmed H. Hassan Al-Marzouqi, Afaf Kamal Eldin, Nabeel Alsabeeha, Thomas E. Adrian, Cesare Stefanini, and et al. 2020. "SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions" Viruses 12, no. 5: 526. https://doi.org/10.3390/v12050526
APA StyleUddin, M., Mustafa, F., Rizvi, T. A., Loney, T., Al Suwaidi, H., Al-Marzouqi, A. H. H., Kamal Eldin, A., Alsabeeha, N., Adrian, T. E., Stefanini, C., Nowotny, N., Alsheikh-Ali, A., & Senok, A. C. (2020). SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses, 12(5), 526. https://doi.org/10.3390/v12050526







