Localization of Tobacco Yellow Dwarf Virus Replication Using the In Plant Activation (INPACT) Expression Platform
Abstract
1. Introduction
2. Materials and Methods
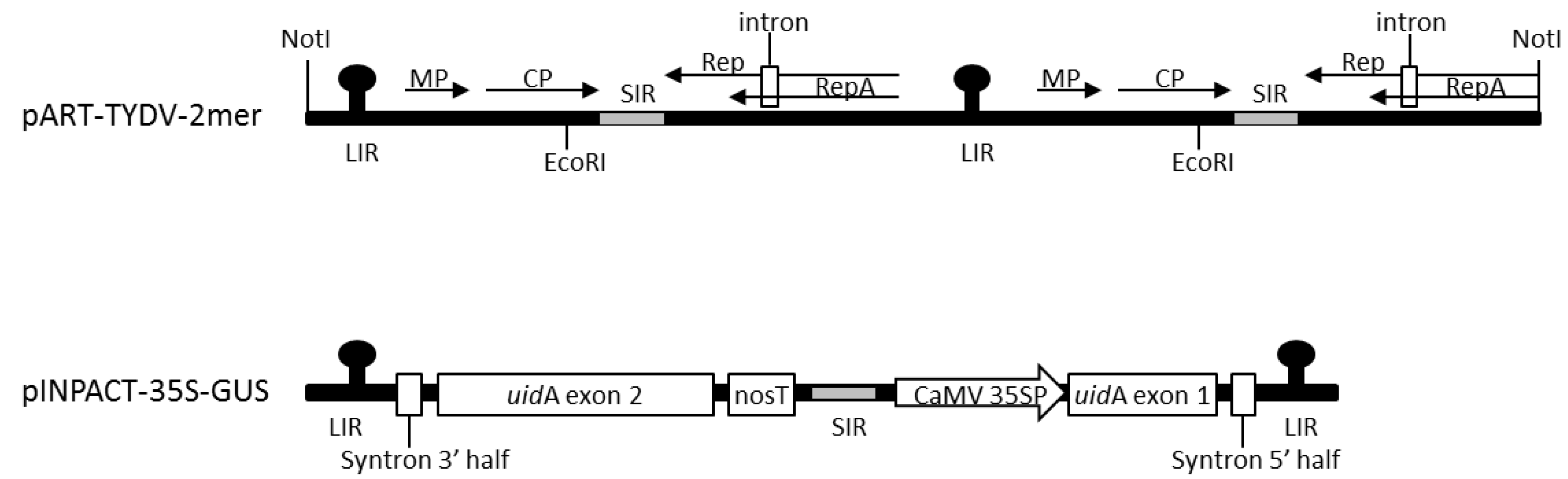
2.1. Vector Construction
2.2. Transformation of Agrobacteria and Tobacco
2.3. Agroinfiltration of Leaves
2.4. GUS Histochemical Staining
2.5. Fixing, Sectioning and Microscopy
2.6. Confirmation of TYDV Infection by PCR
3. Results
3.1. Agroinfiltration of an Infectious Clone to Establish TYDV Infection
3.2. Generation of Transgenic Lines Containing the pINPACT-35S-GUS Expression Cassette
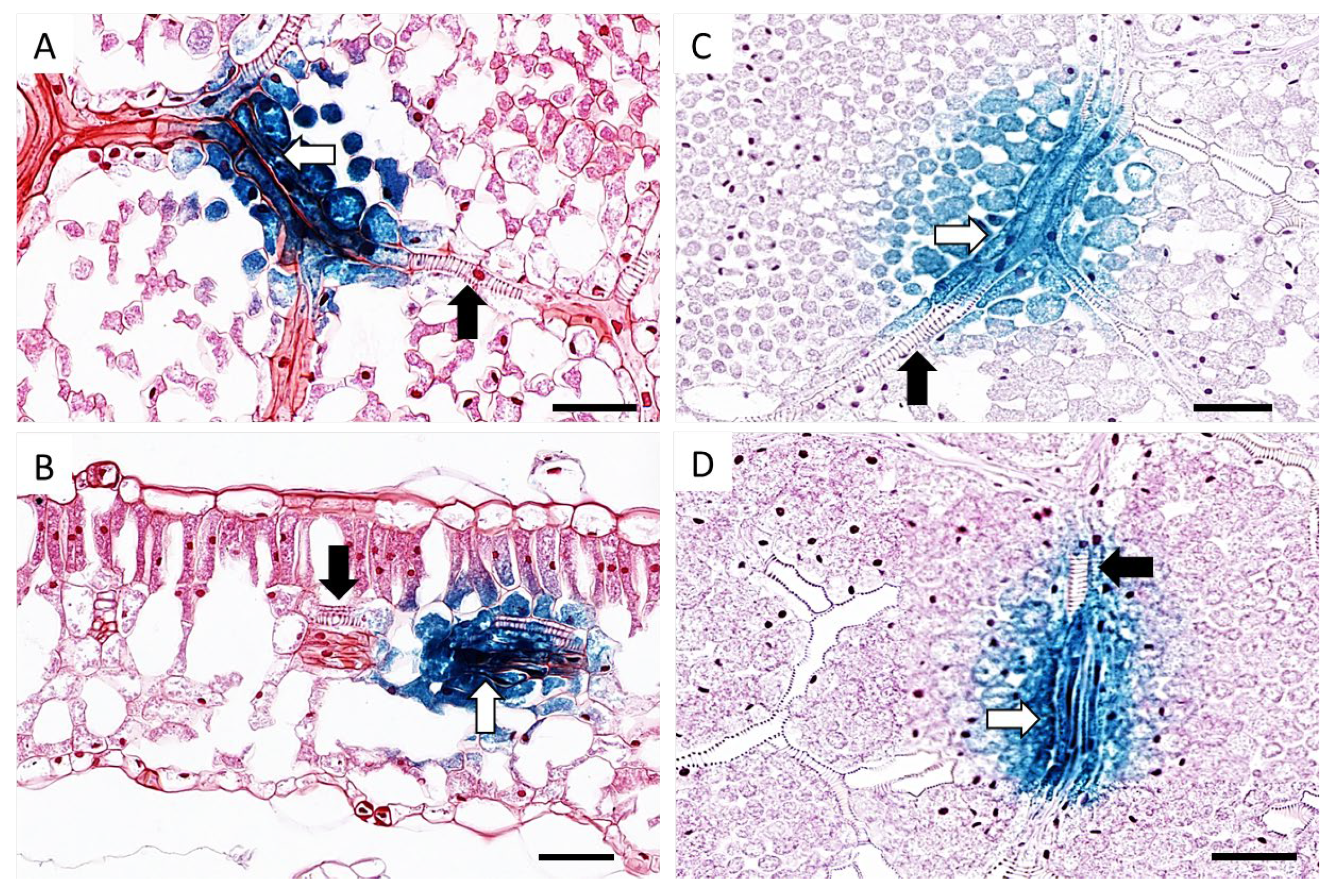
3.3. Cellular Localisation of TYDV Replication
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for Plant DNA Replication, Transcription, and Cell Cycle Regulation. CRC. Crit. Rev. Plant. Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Gutierrez, C. Geminivirus DNA replication. Cell. Mol. Life Sci. 1999, 56, 313–329. [Google Scholar] [CrossRef]
- Alves-Júnior, M.; Alfenas-Zerbini, P.; Andrade, E.C.; Esposito, D.A.; Silva, F.N.; Cláudia, A.; Da Cruz, F.; Ventrella, M.C.; Otoni, W.C.; Murilo Zerbini, F. Synergism and negative interference during co-infection of tomato and Nicotiana benthamiana with two bipartite begomoviruses. Virology 2009, 387, 257–266. [Google Scholar] [CrossRef]
- Bass, H.W.; Nagar, S.; Hanley-Bowdoin, L.; Robertson, D. Chromosome condensation induced by geminivirus infection of mature plant cells. J. Cell Sci. 2000, 113, 1149–1160. [Google Scholar]
- Esau, K. Virus-Like Particles in Nuclei of Phloem Cells in Spinach Leaves Infected with the Curly Top Virus 1. J. Ultrastruct. Res. 1977, 61, 78–88. [Google Scholar] [CrossRef]
- Horns, T.; Jeske, H. Localization of abutilon mosaic virus (AbMV) DNA within leaf tissue by in situ hybridization. Virology 1991, 181, 580–588. [Google Scholar] [CrossRef]
- Lucy, A.P.; Boulton, M.I.; Davies, J.W.; Maule, A.J. Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant.-Microbe. Interact. 1996, 9, 22–31. [Google Scholar] [CrossRef]
- Morilla, G.; Krenz, B.; Jeske, H.; Bejarano, E.R.; Wege, C. Tête à Tête of Tomato Yellow Leaf Curl Virus and Tomato Yellow Leaf Curl Sardinia Virus in Single Nuclei. J. Virol. 2004, 78, 10715–10723. [Google Scholar] [CrossRef] [PubMed]
- Morra, M.R.; Petty, I.T.D. Tissue specificity of geminivirus infection is genetically determined. Plant. Cell 2000, 12, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Nagar, S.; Pedersen, T.J.; Carrick, K.M.; Hanley-Bowdoin, L.; Robertson, D. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant. Cell 1995, 7, 705–719. [Google Scholar] [PubMed]
- Rushing, A.E.; Sunter, G.; Gardiner, W.; Dute, R.; Bisaro, D. Ultrastructural Aspect of Tomato Golden Mosaic Virus Infection in Tobacco. Phytopathology 1987, 77, 1231–1236. [Google Scholar] [CrossRef]
- Sanderfoot, A.A.; Ingham, D.J.; Lazarowitz, S.G. A viral movement protein as a nuclear shuttle: The geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant. Physiol. 1996, 110, 23–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sudarshana, M.R.; Wang, H.L.; Lucas, W.J.; Gilbertson, R.L. Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant.-Microbe Interact. 1998, 11, 277–291. [Google Scholar] [CrossRef][Green Version]
- Rasheed, M.S.; Selth, L.A.; Koltunow, A.M.G.; Randles, J.W.; Rezaian, M.A. Single-stranded DNA of Tomato leaf curl virus accumulates in the cytoplasm of phloem cells. Virology 2006, 348, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Nagar, S.; Hanley-Bowdoin, L.; Robertson, D. Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant. Cell 2002, 14, 2995–3007. [Google Scholar] [CrossRef] [PubMed]
- Morilla, G.; Castillo, A.G.; Preiss, W.; Jeske, H.; Bejarano, E.R. A Versatile Transreplication-Based System To Identify Cellular Proteins Involved in Geminivirus Replication. J. Virol. 2006, 80, 3624–3633. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Deuschle, K.; Deigner, T.; Unseld, S.; Kepp, G.; Wege, C.; Kleinow, T.; Jeske, H. Early Function of the Abutilon Mosaic Virus AC2 Gene as a Replication Brake. J. Virol. 2015, 89, 3683–3699. [Google Scholar] [CrossRef][Green Version]
- Rothenstein, D.; Krenz, B.; Selchow, O.; Jeske, H. Tissue and cell tropism of Indian cassava mosaic virus (ICMV) and its AV2 (precoat) gene product. Virology 2007, 359, 137–145. [Google Scholar] [CrossRef]
- Morris, B.A.M.; Richardson, K.A.; Haley, A.; Zhan, X.; Thomas, J.E. The nucleotide sequence of the infectious cloned DNA component of tobacco yellow dwarf virus reveals features of geminiviruses infecting monocotyledonous plants. Virology 1992, 187, 633–642. [Google Scholar] [CrossRef]
- Borah, B.K.; Zarreen, F.; Baruah, G.; Dasgupta, I. Insights into the control of geminiviral promoters. Virology 2016, 495, 101–111. [Google Scholar] [CrossRef]
- Gutierrez, C.; Ramirez-Parra, E.; Castellano, M.M.; Sanz-Burgos, A.P.; Luque, A.; Missich, R. Geminivirus DNA replication and cell cycle interactions. In Proceedings of the Veterinary Microbiology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 98, pp. 111–119. [Google Scholar]
- Helson, G.A.H. Yellow dwarf of tobacco in Australia V. Transmission by Orosius argentatus (Evans) to some alternative host plants. Aust. J. Agric. Res. 1950, 1, 144–147. [Google Scholar]
- Thomas, J.E.; Bowyer, J.W. Properties of Tobacco Yellow Dwarf and Bean Summer Death Viruses. Phytopathology 1980, 70, 214–217. [Google Scholar] [CrossRef]
- Van Rijswijk, B.; Rodoni, B.C.; Revill, P.A.; Thomas, J.E.; Moran, J.R.; Harding, R.M. Analysis of variability in partial sequences of genomes of Tobacco yellow dwarf virus isolates. Australas. Plant. Pathol. 2004, 33, 367–370. [Google Scholar] [CrossRef]
- Trębicki, P.; Harding, R.M.; Rodoni, B.; Baxter, G.; Powell, K.S. Vectors and alternative hosts of Tobacco yellow dwarf virus in southeastern Australia. Ann. Appl. Biol. 2010, 157, 13–24. [Google Scholar] [CrossRef]
- Hadfield, J.; Thomas, J.E.; Schwinghamer, M.W.; Kraberger, S.; Stainton, D.; Dayaram, A.; Parry, J.N.; Pande, D.; Martin, D.P.; Varsani, A. Molecular characterisation of dicot-infecting mastreviruses from Australia. Virus Res. 2012, 166, 13–22. [Google Scholar] [CrossRef]
- Ghauri, M.S.K. Revision of the genus Orosius Distant (Homoptera: Cicadelloidea). Bull. Br. Museum. Nat. Hist. Entomol. 1966, 18, 231–252. [Google Scholar]
- Atkinson, R.G.; Bieleski, L.R.F.; Gleave, A.P.; Janssen, B.J.; Morris, B.A.M. Post-transcriptional silencing of chalcone synthase in Petunia using a geminivirus-based episomal vector. Plant. J. 1998, 15, 593–604. [Google Scholar] [CrossRef]
- Dugdale, B.; Kato, M.; Deo, P.; Plan, M.; Harrison, M.; Lloyd, R.; Walsh, T.; Harding, R.; Dale, J. Production of human vitronectin in Nicotiana benthamiana using the INPACT hyperexpression platform. Plant. Biotechnol. J. 2018, 16, 394–403. [Google Scholar] [CrossRef]
- Dugdale, B.; Mortimer, C.L.; Kato, M.; James, T.A.; Harding, R.M.; Dale, J.L. In plant activation: An inducible, hyperexpression platformfor recombinant protein production in plants. Plant Cell 2013, 25, 2429–2443. [Google Scholar] [CrossRef]
- Dugdale, B.; Mortimer, C.L.; Kato, M.; James, T.A.; Harding, R.M.; Dale, J.L. Design and construction of an in-plant activation cassette for transgene expression and recombinant protein production in plants. Nat. Protoc. 2014, 9, 1010–1027. [Google Scholar] [CrossRef]
- Stewart, C.N.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 1993, 14, 748–751. [Google Scholar] [PubMed]
- Lin, J.J. Electrotransformation of Agrobacterium. Methods Mol. Biol. 1995, 47, 171–178. [Google Scholar] [PubMed]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A Simple and General Method for Transferring Genes into Plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant. Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant. Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Thomas, J.E.; Bowyer, J.W. Tobacco yellow dwarf virus. C. Descr. Plant. Viruses 1984, 4, 278. [Google Scholar]
- Needham, P.D.; Atkinson, R.G.; Morris, B.A.M.; Gardner, R.C.; Gleave, A.P. GUS expression patterns from a tobacco yellow dwarf virus-based episomal vector. Plant. Cell Rep. 1998, 17, 631–639. [Google Scholar] [CrossRef]
- Diamos, A.G.; Mason, H.S. Modifying the Replication of Geminiviral Vectors Reduces Cell Death and Enhances Expression of Biopharmaceutical Proteins in Nicotiana benthamiana Leaves. Front. Plant. Sci. 2018, 9, 1974. [Google Scholar] [CrossRef]
- Wydro, M.; Kozubek, E.; Lehmann, P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 2006, 53, 289–298. [Google Scholar] [CrossRef]
- Wroblewski, T.; Tomczak, A.; Michelmore, R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant. Biotechnol. J. 2005, 3, 259–273. [Google Scholar] [CrossRef]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Gilbertson, R.L.; Lucas, W.J. Spatial and Temporal Distribution of Bean Dwarf Mosaic Geminivirus in Phaseolus vulgaris and Nicotiana benthamiana. Phytopathology 1996, 86, 1204–1214. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Dugdale, B.; Martin, D.P.; Varsani, A.; Lakay, F.M.; Bezuidenhout, M.E.; Monjane, A.L.; Thomson, J.A.; Dale, J.; Rybicki, E.P. Inducible resistance to maize streak virus. PLoS ONE 2014, 9, e105932. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, M.; Harding, R.; Dale, J.; Dugdale, B. Localization of Tobacco Yellow Dwarf Virus Replication Using the In Plant Activation (INPACT) Expression Platform. Viruses 2020, 12, 688. https://doi.org/10.3390/v12060688
Kato M, Harding R, Dale J, Dugdale B. Localization of Tobacco Yellow Dwarf Virus Replication Using the In Plant Activation (INPACT) Expression Platform. Viruses. 2020; 12(6):688. https://doi.org/10.3390/v12060688
Chicago/Turabian StyleKato, Maiko, Robert Harding, James Dale, and Benjamin Dugdale. 2020. "Localization of Tobacco Yellow Dwarf Virus Replication Using the In Plant Activation (INPACT) Expression Platform" Viruses 12, no. 6: 688. https://doi.org/10.3390/v12060688
APA StyleKato, M., Harding, R., Dale, J., & Dugdale, B. (2020). Localization of Tobacco Yellow Dwarf Virus Replication Using the In Plant Activation (INPACT) Expression Platform. Viruses, 12(6), 688. https://doi.org/10.3390/v12060688




