Yellow Fever Virus Down-Regulates mRNA Expression of SOCS1 in the Initial Phase of Infection in Human Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Experimental Design
2.3. Plaque Assay
2.4. RT-qPCR
2.5. Western Blot Analysis
2.6. Statistical Analysis
3. Results
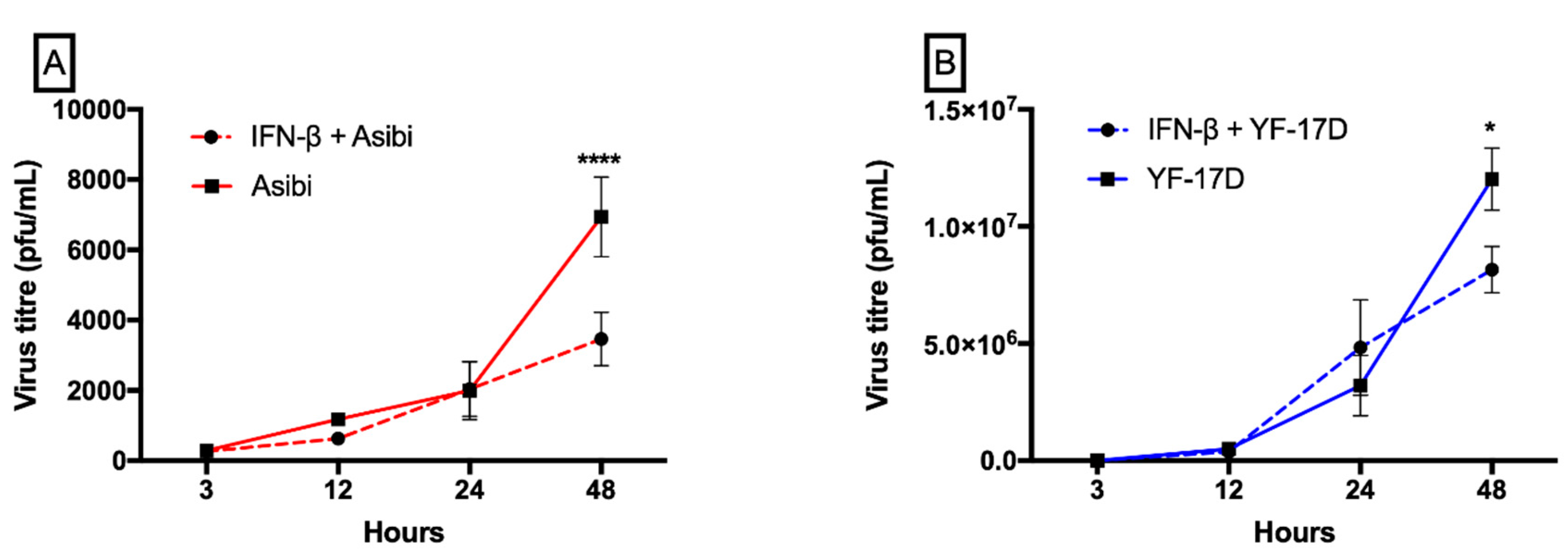
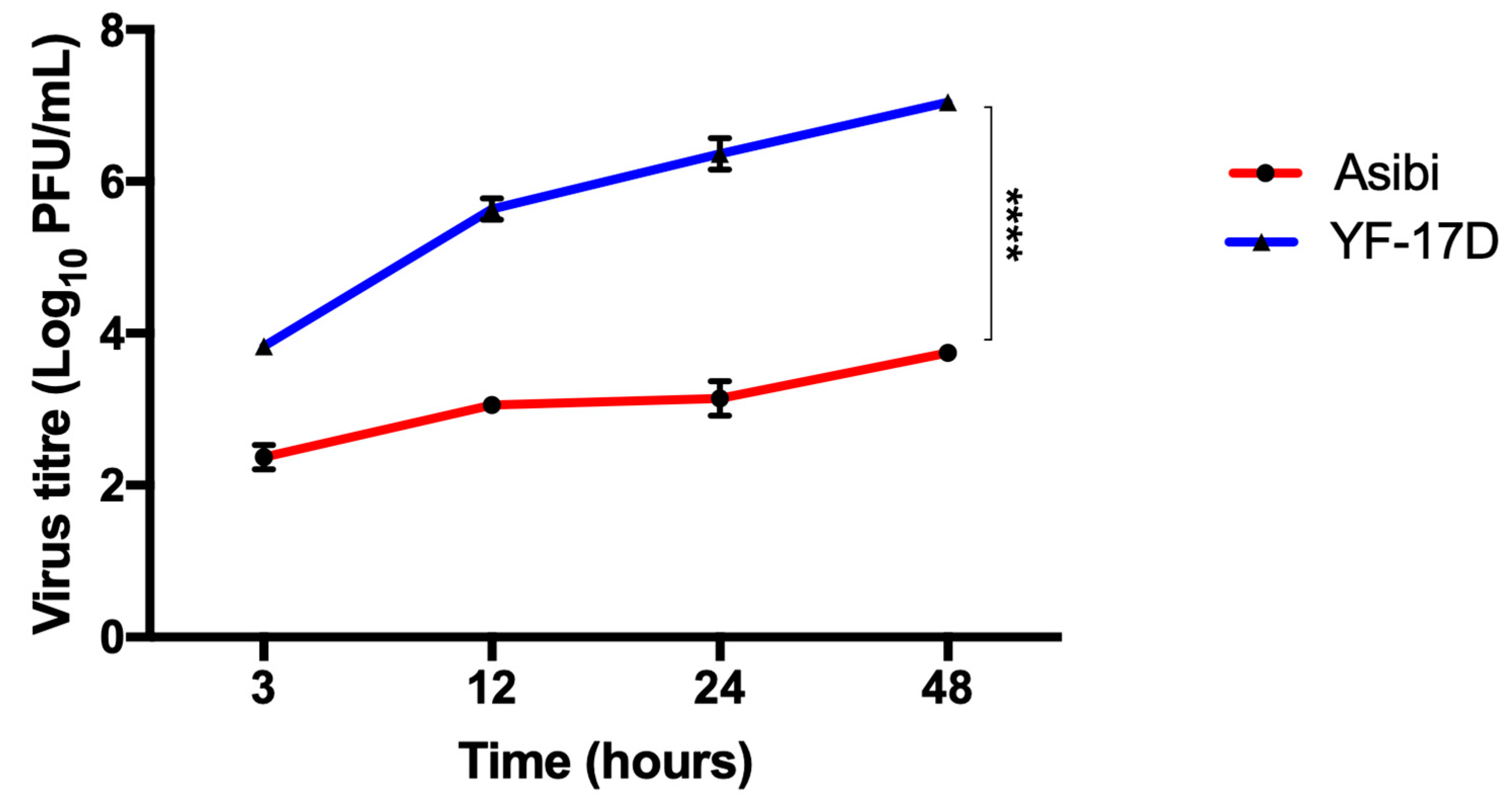
3.1. IFN-β Inhibits both YFV Strains at Later Time Points whilst YF-17D Replicates More Efficiently
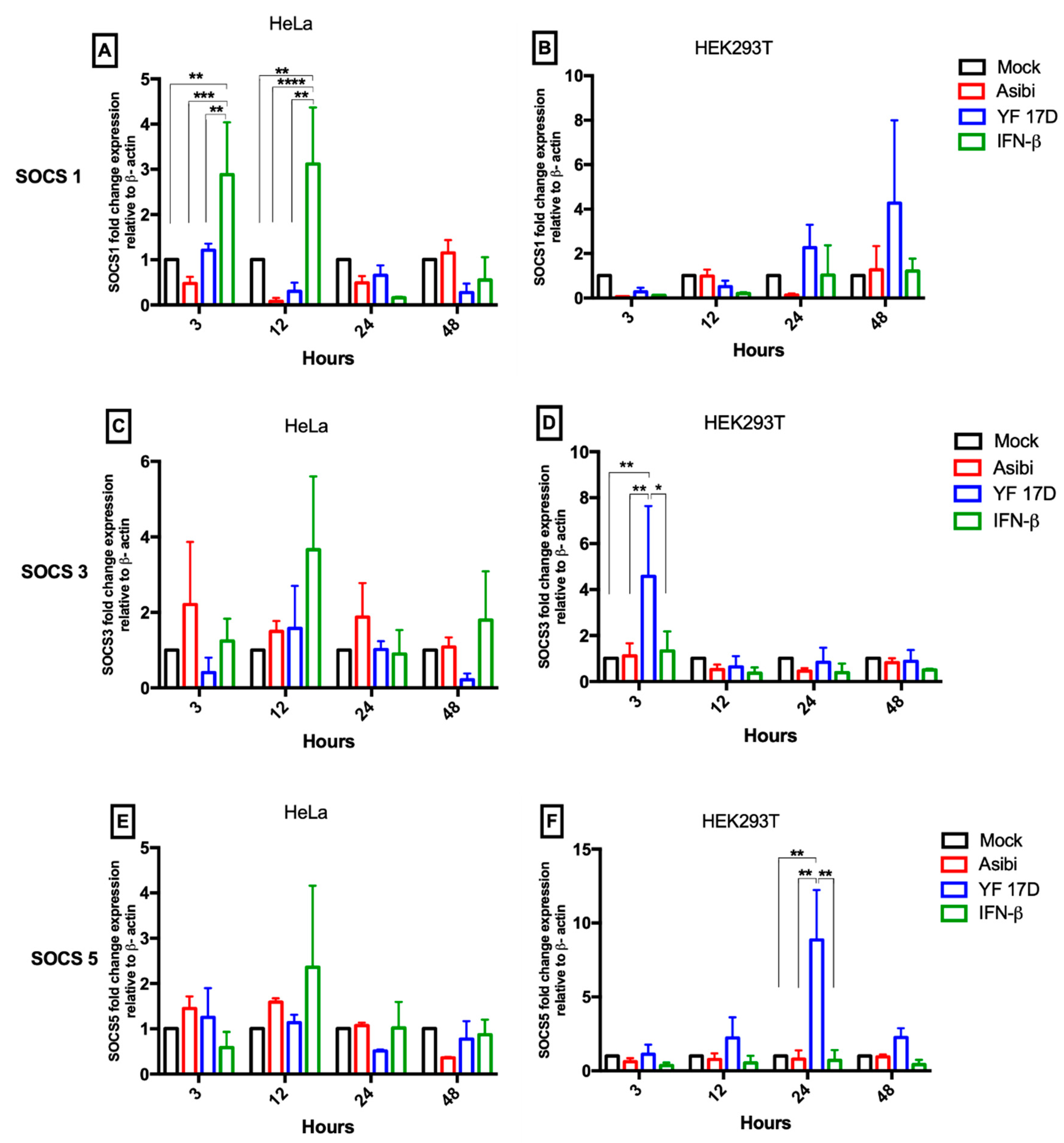
3.2. Expression Dynamics of SOCS Genes
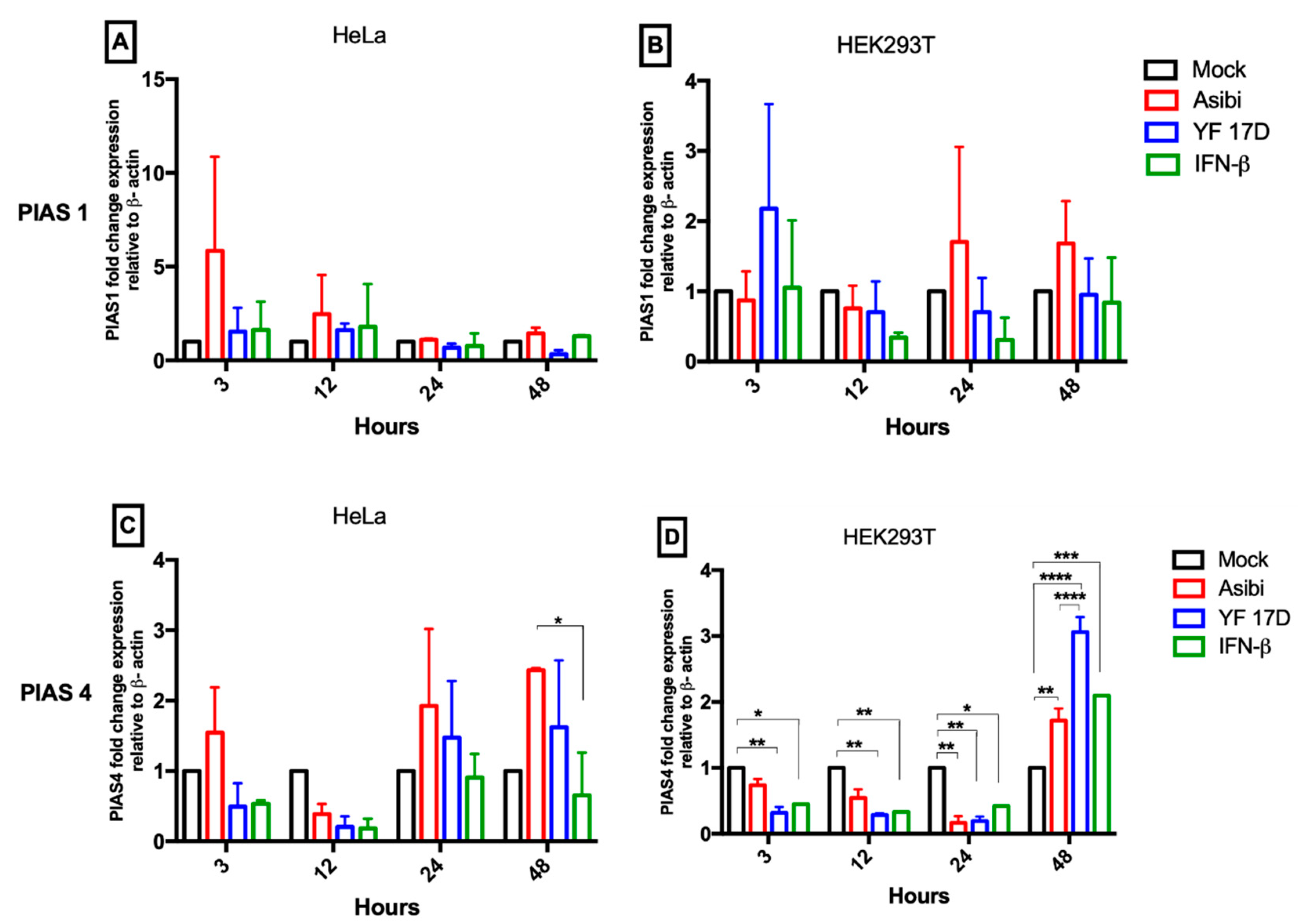
3.3. Expression Dynamics of PIAS Genes
3.4. Expression Dynamics of Antiviral Molecules
3.5. Effect of IFN-β on YFV Induction of Antiviral Molecules
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, J.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-Viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Naka, T.; Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007, 7, 454–465. [Google Scholar] [CrossRef]
- Shuai, K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006, 16, 196–202. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, L.N.; Benveniste, E.N. Viral exploitation of host SOCS protein functions. J. Virol. 2011, 85, 1912–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotaja, N.; Karvonen, U.; Janne, O.A.; Palvimo, J.J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002, 22, 5222–5234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, L.N.; Qin, H.; Muldowney, M.T.; Yanagisawa, L.L.; Kutsch, O.; Clements, J.E.; Benveniste, E.N. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J. Immunol. 2010, 185, 2393–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-Jimenez, T.; Millan-Perez Pena, L.; Flores-Mendoza, L.; Sedeno-Monge, V.; Santos-Lopez, G.; Rosas-Murrieta, N.; Reyes-Carmona, S.; Teran-Cabanillas, E.; Hernandez, J.; Herrera-Camacho, I.; et al. Upregulation of the suppressors of cytokine signaling 1 and 3 is associated with arrest of phosphorylated-stat1 nuclear importation and reduced innate response in denguevirus-infected macrophages. Viral Immunol. 2016, 29, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Palma-Ocampo, H.K.; Flores-Alonso, J.C.; Vallejo-Ruiz, V.; Reyes-Leyva, J.; Flores-Mendoza, L.; Herrera-Camacho, I.; Rosas-Murrieta, N.H.; Santos-López, G. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol. J. 2015, 12, 150. [Google Scholar] [CrossRef] [Green Version]
- Kundu, K.; Dutta, K.; Nazmi, A.; Basu, A. Japanese encephalitis virus infection modulates the expression of suppressors of cytokine signaling (SOCS) in macrophages: Implications for the hosts’ innate immune response. Cell. Immunol. 2013, 285, 100–110. [Google Scholar] [CrossRef]
- Faria, N.R.; Kraemer, M.U.G.; Hill, S.C.; Goes de Jesus, J.; Aguiar, R.S.; Iani, F.C.M.; Xavier, J.; Quick, J.; du Plessis, L.; Dellicour, S.; et al. Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science 2018, 361, 894–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theiler, M.; Smith, H.H. The use of yellow fever virus modified by in vitro cultivation for human immunization. J. Exp. Med. 1937, 65, 787–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, A.S.; Barrett, A.D. Current status and future prospects of yellow fever vaccines. Expert Rev. Vaccines 2015, 14, 1479–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.E. Yellow fever vaccine-associated viscerotropic disease: Current perspectives. Drug Des. Dev. Ther. 2016, 10, 3345–3353. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO position on the use of fractional doses—June 2017, addendum to vaccines and vaccination against yellow fever WHO: Position paper—June 2013. Vaccine 2017, 35, 5751–5752. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Yellow fever vaccine. Expert Rev. Vaccines 2005, 4, 553–574. [Google Scholar] [CrossRef]
- Monath, T.P.; Lee, C.K.; Julander, J.G.; Brown, A.; Beasley, D.W.; Watts, D.M.; Hayman, E.; Guertin, P.; Makowiecki, J.; Crowell, J.; et al. Inactivated yellow fever 17D vaccine: Development and nonclinical safety, immunogenicity and protective activity. Vaccine 2010, 28, 3827–3840. [Google Scholar] [CrossRef]
- Freire, M.S.; Mann, G.F.; Marchevsky, R.S.; Yamamura, A.M.; Almeida, L.F.; Jabor, A.V.; Malachias, J.M.; Coutinho, E.S.; Galler, R. Production of yellow fever 17DD vaccine virus in primary culture of chicken embryo fibroblasts: Yields, thermo and genetic stability, attenuation and immunogenicity. Vaccine 2005, 23, 2501–2512. [Google Scholar] [CrossRef] [Green Version]
- Beck, A.; Tesh, R.B.; Wood, T.G.; Widen, S.G.; Ryman, K.D.; Barrett, A.D. Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. J. Infect. Dis. 2014, 209, 334–344. [Google Scholar] [CrossRef]
- Fernandez-Garcia, M.D.; Meertens, L.; Chazal, M.; Hafirassou, M.L.; Dejarnac, O.; Zamborlini, A.; Despres, P.; Sauvonnet, N.; Arenzana-Seisdedos, F.; Jouvenet, N.; et al. Vaccine and wild-type strains of yellow fever virus engage distinct entry mechanisms and differentially stimulate antiviral immune responses. mBio 2016, 7, e01956-15. [Google Scholar] [CrossRef] [Green Version]
- Diamond, M.S.; Harris, E. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 2001, 289, 297–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, B.C.; Fredericksen, B.L.; Samuel, M.A.; Mock, R.E.; Mason, P.W.; Diamond, M.S.; Gale, M., Jr. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 2006, 80, 9424–9434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, Y.; McArthur, M.A.; Cohen, M.; Jahrling, P.B.; Janosko, K.B.; Josleyn, N.; Kang, K.; Zhang, T.; Holbrook, M.R. Characterization of yellow fever virus infection of human and non-human primate antigen presenting cells and their interaction with CD4+ T Cells. PLoS Negl. Trop. Dis. 2016, 10, e0004709. [Google Scholar] [CrossRef] [PubMed]
- Akondy, R.S.; Johnson, P.L.; Nakaya, H.I.; Edupuganti, S.; Mulligan, M.J.; Lawson, B.; Miller, J.D.; Pulendran, B.; Antia, R.; Ahmed, R. Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc. Natl. Acad. Sci. USA 2015, 112, 3050–3055. [Google Scholar] [CrossRef] [Green Version]
- Watson, A.M.; Lam, L.K.M.; Klimstra, W.B.; Ryman, K.D. The 17D-204 vaccine strain-induced protection against virulent yellow fever virus is mediated by humoral immunity and CD4+ but not CD8+ T Cells. PLoS Pathog. 2016, 12, e1005786. [Google Scholar] [CrossRef]
- Querec, T.D.; Akondy, R.S.; Lee, E.K.; Cao, W.; Nakaya, H.I.; Teuwen, D.; Pirani, A.; Gernert, K.; Deng, J.; Marzolf, B.; et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009, 10, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Seong, R.K.; Lee, J.K.; Shin, O.S. Zika virus-induction of the suppressor of cytokine signaling 1/3 contributes to the modulation of viral replication. Pathogens (Basel, Switzerland) 2020, 9, 163. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Hou, M.; Liu, X.; Li, Z.; Yang, Y.; Zhang, W. Induction of SOCS expression by EV71 infection promotes EV71 replication. BioMed Res. Int. 2020, 2020, 2430640. [Google Scholar] [CrossRef] [Green Version]
- Steffensen, M.A.; Fenger, C.; Christensen, J.E.; Jørgensen, C.K.; Bassi, M.R.; Christensen, J.P.; Finsen, B.; Thomsen, A.R. Suppressors of cytokine signaling 1 and 3 are upregulated in brain resident cells in response to virus-induced inflammation of the central nervous system via at least two distinctive pathways. J. Virol. 2014, 88, 14090–14104. [Google Scholar] [CrossRef] [Green Version]
- Souma, Y.; Nishida, T.; Serada, S.; Iwahori, K.; Takahashi, T.; Fujimoto, M.; Ripley, B.; Nakajima, K.; Miyazaki, Y.; Mori, M.; et al. Antiproliferative effect of SOCS-1 through the suppression of STAT3 and p38 MAPK activation in gastric cancer cells. Int. J. Cancer 2012, 131, 1287–1296. [Google Scholar] [CrossRef]
- Laurent-Rolle, M.; Morrison, J.; Rajsbaum, R.; Macleod, J.M.; Pisanelli, G.; Pham, A.; Ayllon, J.; Miorin, L.; Martinez-Romero, C.; tenOever, B.R.; et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 2014, 16, 314–327. [Google Scholar] [CrossRef] [Green Version]
- ter Meulen, J.; Sakho, M.; Koulemou, K.; Magassouba, N.F.; Bah, A.; Preiser, W.; Daffis, S.; Klewitz, C.; Bae, H.-G.; Niedrig, M.; et al. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J. Infect. Dis. 2004, 190, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Yokosawa, N.; Okabayashi, T.; Suzutani, T.; Fujii, N. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 confers efficient viral replication. Virology 2005, 338, 173–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Wang, S.; Sun, F.; Zheng, G.; Wu, T.; Du, Y.; Zhang, S.; Qian, J.; Sun, R. Inhibition of murine herpesvirus-68 replication by IFN-gamma in macrophages is counteracted by the induction of SOCS1 expression. PLoS Pathog. 2018, 14, e1007202. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Magri, A.; Hill, M.; Lai, A.G.; Kumar, A.; Rambhatla, S.B.; Donald, C.L.; Lopez-Clavijo, A.F.; Rudge, S.; Pinnick, K.; et al. The circadian clock components BMAL1 and REV-ERBalpha regulate flavivirus replication. Nat. Commun. 2019, 10, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | 5′-Sequence-3′ |
|---|---|
| IFN-α F | TCGCCCTTTGCTTTACTGAT |
| IFN-α R | GGGTCTCAGGGAGATCACAG |
| MxA F | CAGTTGAGGGCAAGGAGTGT |
| MxA R | ATGCCAGGAACCCACATACG |
| OAS1 F | AGCAACAGTGCAGACGATGA |
| OAS1 R | TTGGCTCTGTGCCTTGAAGT |
| PIAS1 F | GCAGACTTGTCCATCCCCAA |
| PIAS1 R | ACTGGGTCAAAGTAAAAGCCT |
| PIAS4 F | CTGGCACTTCCCATACCTGT |
| PIAS4 R | GGGATGGGAGAAGGACTAGC |
| β-Actin F | ATGATATCGCCGCGCTCGTC |
| β-Actin R | CGCTCGGTGAGGATCTTCA |
| SOCS-1 F | AGACCCCTTCTCACCTCTTG |
| SOCS-1 R | CTGCACAGCAGAAAATAAAGC |
| SOCS-3 F | TCCCCCCAGAAGAGCCTATTAC |
| SOCS-3 R | TCCGACAGAGATGCTGAAGAGTG |
| SOCS-5 F | AGTCAAAGCCTCTCTTTTCC |
| SOCS-5 R | ACTGAACCTGACCGTACACATTTTTGGGCTAAATCTGA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakass, M.B.; Franco, D.; Quaye, O. Yellow Fever Virus Down-Regulates mRNA Expression of SOCS1 in the Initial Phase of Infection in Human Cell Lines. Viruses 2020, 12, 802. https://doi.org/10.3390/v12080802
Yakass MB, Franco D, Quaye O. Yellow Fever Virus Down-Regulates mRNA Expression of SOCS1 in the Initial Phase of Infection in Human Cell Lines. Viruses. 2020; 12(8):802. https://doi.org/10.3390/v12080802
Chicago/Turabian StyleYakass, Michael B., David Franco, and Osbourne Quaye. 2020. "Yellow Fever Virus Down-Regulates mRNA Expression of SOCS1 in the Initial Phase of Infection in Human Cell Lines" Viruses 12, no. 8: 802. https://doi.org/10.3390/v12080802






