One of the Triple Poly(A) Signals in the M112-113 Gene Is Important and Sufficient for Stabilizing the M112-113 mRNA and the Replication of Murine Cytomegalovirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Culture
2.2. Antibodies
2.3. Immunofluorescent Assay (IFA)
2.4. Plasmids and Molecular Cloning
2.5. Bacterial Artificial Chromosome (BAC) Mutagenesis to Generate MCMVs with Mutated E1 Poly(A)
2.6. RNA Isolation, Treatment with DNase I (RNase Free), and Real-Time RT-PCR
2.7. Immunoblot Analysis
2.8. Plaque Formation Unit (PFU) Assay
2.9. M112-113 mRNA Stability Assay
3. Results
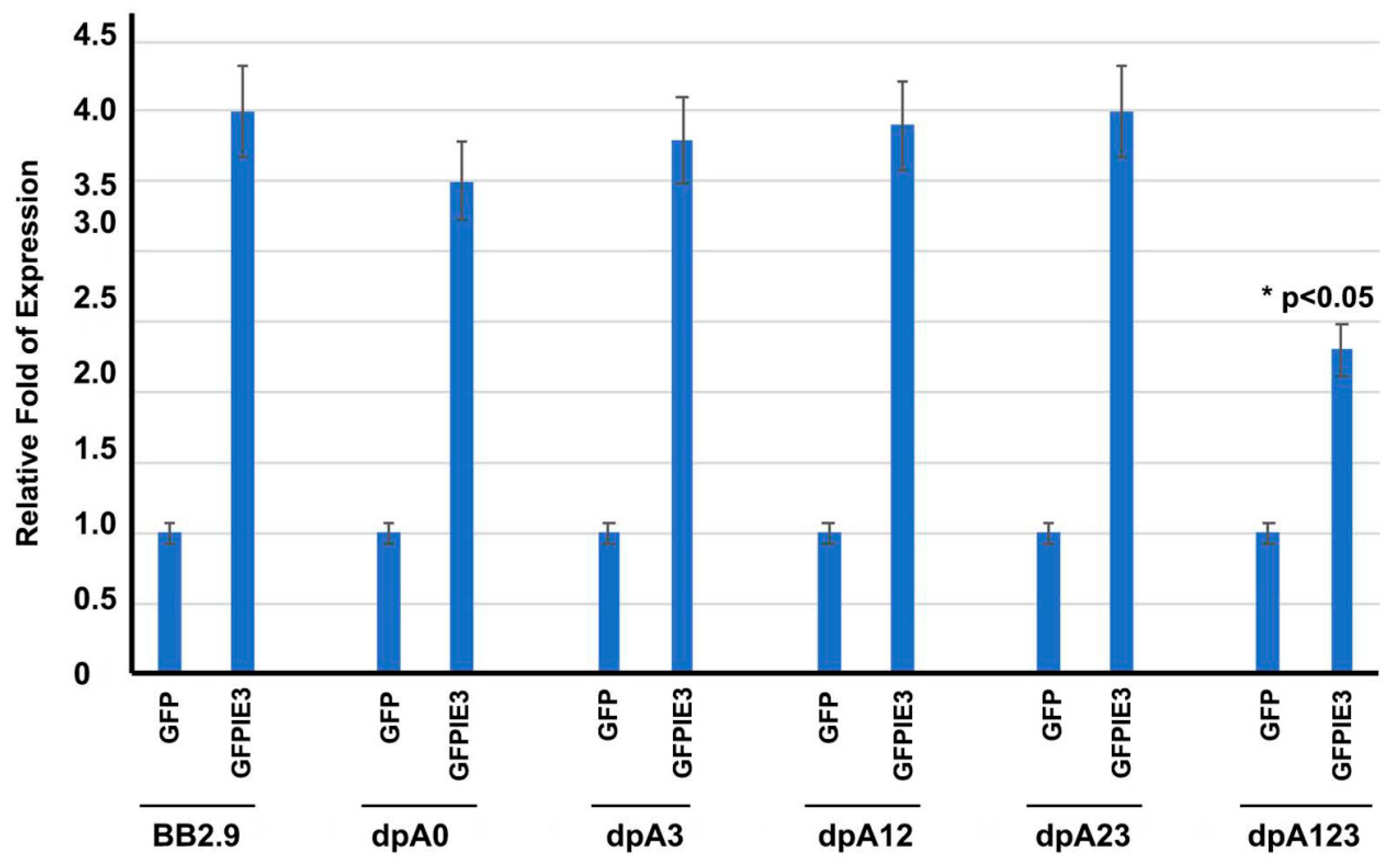
3.1. One of the POLY(A) SIGNALS Is Enough for IE3 to Activate M112-113 Gene Expression
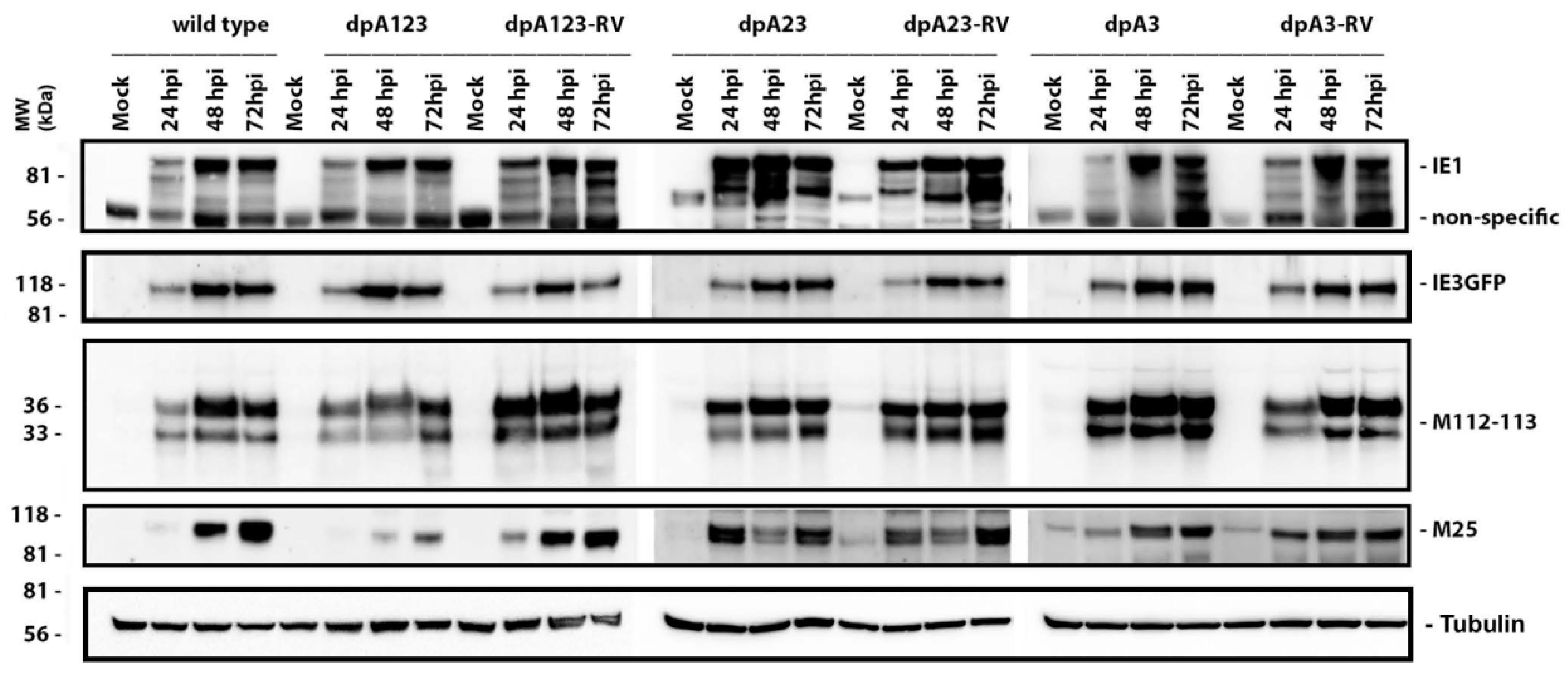
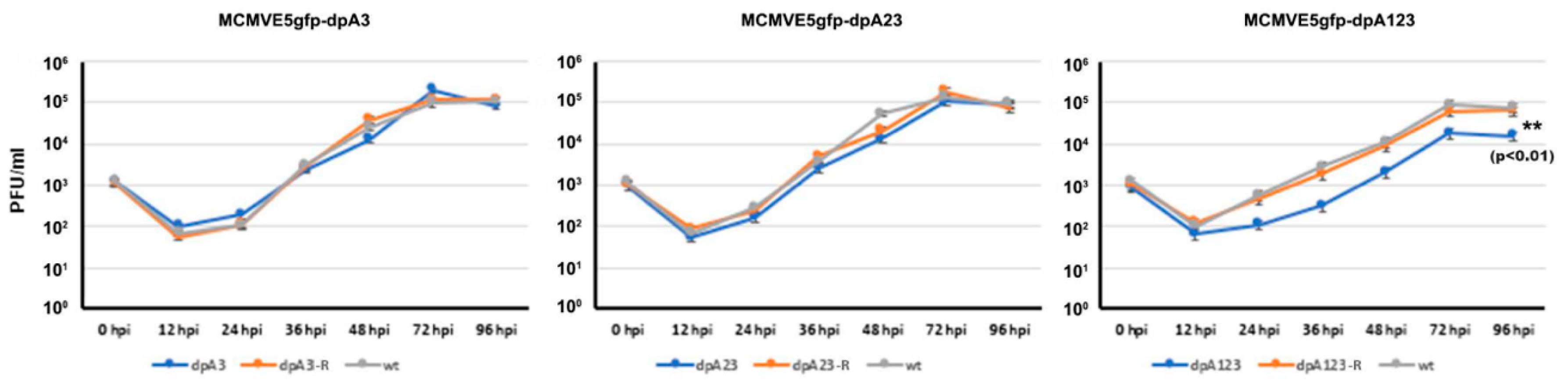
3.2. Deletion of the M112-113 Poly(A) Signal Cluster Lessens MCMV Replication
3.3. One M112-113 Poly(A) Signal Is Sufficient for MCMV to Productively Replicate
3.4. One M112-113 Poly(A) Signal Is Needed and Sufficient to Stabilize the M112-113 mRNA
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Reddehase, M.J.; Simon, C.O.; Seckert, C.K.; Lemmermann, N.; Grzimek, N.K. Murine model of cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 2008, 325, 315–331. [Google Scholar] [PubMed]
- Mocarski, E.S., Jr.; Shenk, T.; Griffins, P.D.; Pass, R.F. Cytomegaloviruses, 5th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Buhler, B.; Keil, G.M.; Weiland, F.; Koszinowski, U.H. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J. Virol. 1990, 64, 1907–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarisky, R.T.; Hayward, G.S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 1996, 70, 7398–7413. [Google Scholar] [CrossRef] [Green Version]
- Pari, G.S.; Anders, D.G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 1993, 67, 6979–6988. [Google Scholar] [CrossRef] [Green Version]
- Arai, Y.; Ishiwata, M.; Baba, S.; Kawasaki, H.; Kosugi, I.; Li, R.Y.; Tsuchida, T.; Miura, K.; Tsutsui, Y. Neuron-specific activation of murine cytomegalovirus early gene e1 promoter in transgenic mice. Am. J. Pathol. 2003, 163, 643–652. [Google Scholar] [CrossRef] [Green Version]
- Wells, R.; Stensland, L.; Vieira, J. The human cytomegalovirus UL112–113 locus can activate the full Kaposi’s sarcoma-associated herpesvirus lytic replication cycle. J. Virol. 2009, 83, 4695–4699. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, U.; Handke, W.; Jurak, I.; Brune, W. Mutations in the M112/M113 coding region facilitate murine cytomegalovirus replication in human cells. J. Virol. 2010, 84, 7994–8006. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Li, L.; Maul, G.G. Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J. Virol. 2005, 79, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Ciocco-Schmitt, G.M.; Karabekian, Z.; Godfrey, E.W.; Stenberg, R.M.; Campbell, A.E.; Kerry, J.A. Identification and characterization of novel murine cytomegalovirus M112–113 (e1) gene products. Virology 2002, 294, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Cizkova, D.; Baird, S.J.E.; Tesikova, J.; Voigt, S.; Ludovit, D.; Pialek, J.; de Bellocq, J.G. Host subspecific viral strains in European house mice: Murine cytomegalovirus in the Eastern (Mus musculus musculus) and Western house mouse (Mus musculus domesticus). Virology 2018, 521, 92–98. [Google Scholar] [CrossRef]
- Schwartz, R.; Helmich, B.; Spector, D.H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 1996, 70, 6955–6966. [Google Scholar] [CrossRef] [Green Version]
- Lang, D.; Gebert, S.; Arlt, H.; Stamminger, T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 1995, 69, 6030–6037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodems, S.M.; Clark, C.L.; Spector, D.H. Separate DNA elements containing ATF/CREB and IE86 binding sites differentially regulate the human cytomegalovirus UL112–113 promoter at early and late times in the infection. J. Virol. 1998, 72, 2697–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arlt, H.; Lang, D.; Gebert, S.; Stamminger, T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 1994, 68, 4117–4125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spector, D.H. Activation and regulation of human cytomegalovirus early genes. Intervirology 1996, 39, 361–377. [Google Scholar] [CrossRef]
- Wen, F.; Armstrong, N.; Hou, W.; Cruz-Cosme, R.; Obwolo, L.A.; Ishizuka, K.; Ullah, H.; Luo, M.H.; Sawa, A.; Tang, Q. Zika virus increases mind bomb 1 levels, causing degradation of pericentriolar material 1 (PCM1) and dispersion of PCM1-containing granules from the centrosome. J. Biol. Chem. 2019, 294, 18742–18755. [Google Scholar] [CrossRef]
- Hengel, H.; Lucin, P.; Jonjic, S.; Ruppert, T.; Koszinowski, U.H. Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J. Virol. 1994, 68, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.A.; Carlson, M.E.; Henry, S.C.; Shanley, J.D. The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology 1999, 262, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Loh, L.C.; Keeler, V.D.; Shanley, J.D. Sequence requirements for the nuclear localization of the murine cytomegalovirus M44 gene product pp50. Virology 1999, 259, 43–59. [Google Scholar] [CrossRef] [Green Version]
- Perez, K.J.; Martinez, F.P.; Cosme-Cruz, R.; Perez-Crespo, N.M.; Tang, Q. A short cis-acting motif in the M112–113 promoter region is essential for IE3 to activate M112–113 gene expression and is important for murine cytomegalovirus replication. J. Virol. 2013, 87, 2639–2647. [Google Scholar] [CrossRef] [Green Version]
- Martinez, F.P.; Cosme, R.S.; Tang, Q. Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J. Gen. Virol. 2010, 91, 2664–2676. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Jonjic, S.; Koszinowski, U.H.; Messerle, M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 1999, 73, 7056–7060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warden, C.; Tang, Q.; Zhu, H. Herpesvirus BACs: Past, present, and future. J. Biomed. Biotechnol. 2011, 2011, 124595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratnadiwakara, M.; Änkö, M. mRNA Stability Assay Using Transcription Inhibition by Actinomycin D in Mouse Pluripotent Stem Cells. Bio-Protocol 2018, 8, e3072. [Google Scholar] [CrossRef]
- Colgan, D.F.; Manley, J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997, 11, 2755–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaudoing, E.; Freier, S.; Wyatt, J.R.; Claverie, J.M.; Gautheret, D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000, 10, 1001–1010. [Google Scholar] [CrossRef] [Green Version]




| E1ClaI | TTATCGATCGCCTCGTCTCGCT |
| PstIe1rev | TGGGGGCTGCAGGCTCAGAAGAAGGAATAC |
| E1PstIdlPA123_Rv | tgggggctgcagACATGGATGTTATGTTGTTAGC |
| E1PstIdlPA23_Rv | tgggggctgcagGATAAAATATTAAAAAATTAA |
| E1PstIdlPA12-Rv | tgggggctgcagCGTGTTTATTGTAACAAATCTC |
| E1PstIdlPA3_Rv | tgggggctgcagGTAACAAATGTGTGGTTTTT |
| E1PstIdlPA0_Rv | tgggggctgcagTGTTTATTGTAACAAATGTG |
| E1polyA_galK_F | TGTGTGTCTCTGTCTACCGGGCCGATGGAGATATTATCCCTGGTCCCCCTCT CCTGTTGACAATTAATCATCGG CA |
| E1polyA_galK_R | TAACCACCGATGAGAATAATATTATTATCTCCCCCACAACCTCCTCCAACTC TCAGCACTGTCCTGCTCCTT |
| E1polyA_F | TGTGTGTCTCTGTCTACCG |
| E1polyAdpA123_R | TAACCACCGATGAGAATAATATTATTATCTCCCCCACAACCTCCTCCAACTC ggg ctg cag aca tgg atg |
| E1polyAdpA23_R | TAACCACCGATGAGAATAATATTATTATCTCCCCCACAACCTCCTCCAACTC ggg ctg cag gat aaa ata |
| E1polyAdpA3_R | TAACCACCGATGAGAATAATATTATTATCTCCCCCACAACCTCCTCCAACTC ggg ctg cag gta aca aat |
| M112-113-Forward | ACT GCA CGA ACG GCG AGG |
| M112-113-Reverse | CAG GAT GAC TTG CAG CAA |
| GAPDH-Forward | GAAGGTGAAGGTCGGAGTC |
| GAPDH-Reverse | GAAGATGGTGATGGGATTTC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-cosme, R.; Armstrong, N.; Tang, Q. One of the Triple Poly(A) Signals in the M112-113 Gene Is Important and Sufficient for Stabilizing the M112-113 mRNA and the Replication of Murine Cytomegalovirus. Viruses 2020, 12, 954. https://doi.org/10.3390/v12090954
Cruz-cosme R, Armstrong N, Tang Q. One of the Triple Poly(A) Signals in the M112-113 Gene Is Important and Sufficient for Stabilizing the M112-113 mRNA and the Replication of Murine Cytomegalovirus. Viruses. 2020; 12(9):954. https://doi.org/10.3390/v12090954
Chicago/Turabian StyleCruz-cosme, Ruth, Najealicka Armstrong, and Qiyi Tang. 2020. "One of the Triple Poly(A) Signals in the M112-113 Gene Is Important and Sufficient for Stabilizing the M112-113 mRNA and the Replication of Murine Cytomegalovirus" Viruses 12, no. 9: 954. https://doi.org/10.3390/v12090954





