Comprehensive Analysis of Codon Usage on Porcine Astrovirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Data
2.2. Phylogenetic Analysis
2.3. Nucleotide Composition
2.4. Effective Number of Codons (ENC)
2.5. Relative Synonymous Codon Usage (RSCU)
2.6. Principal Component Analysis (PCA)
2.7. ENC-Plot Analysis
2.8. Neutrality Plot Analysis
2.9. Parity Rule 2 (PR2) Analysis
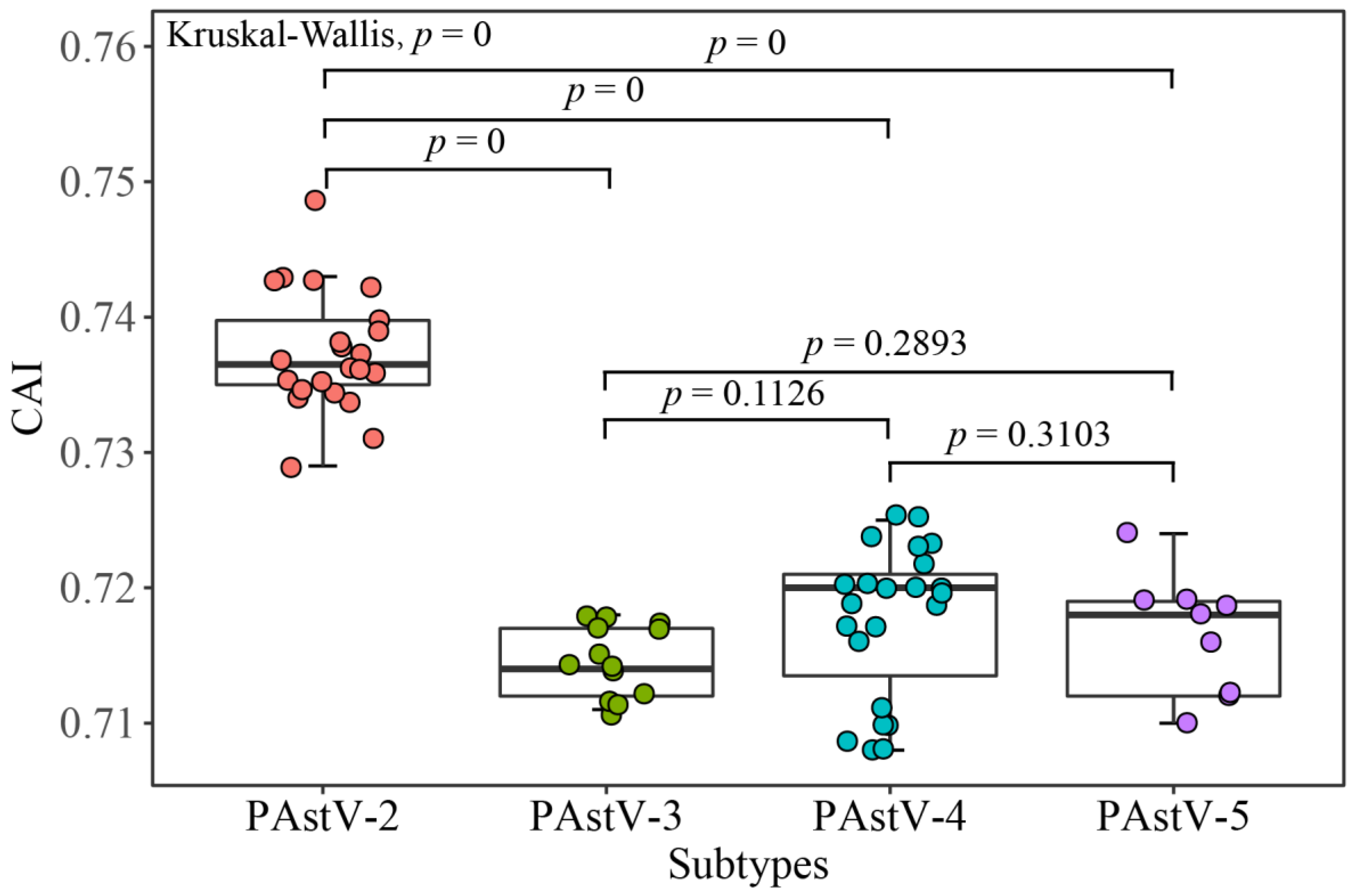
2.10. Codon Adaptation Index (CAI) Analysis
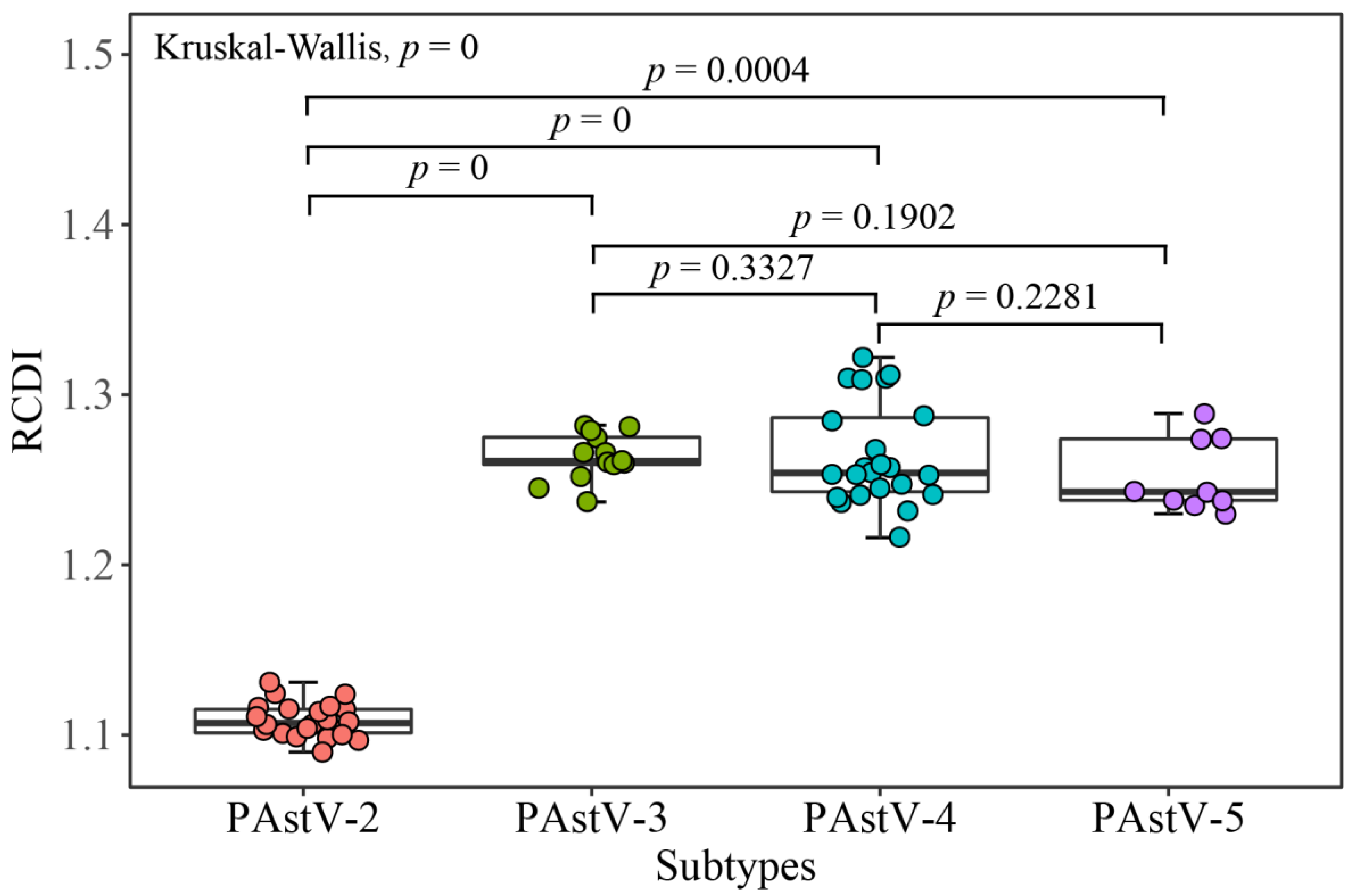
2.11. Relative Codon Deoptimization Index (RCDI)
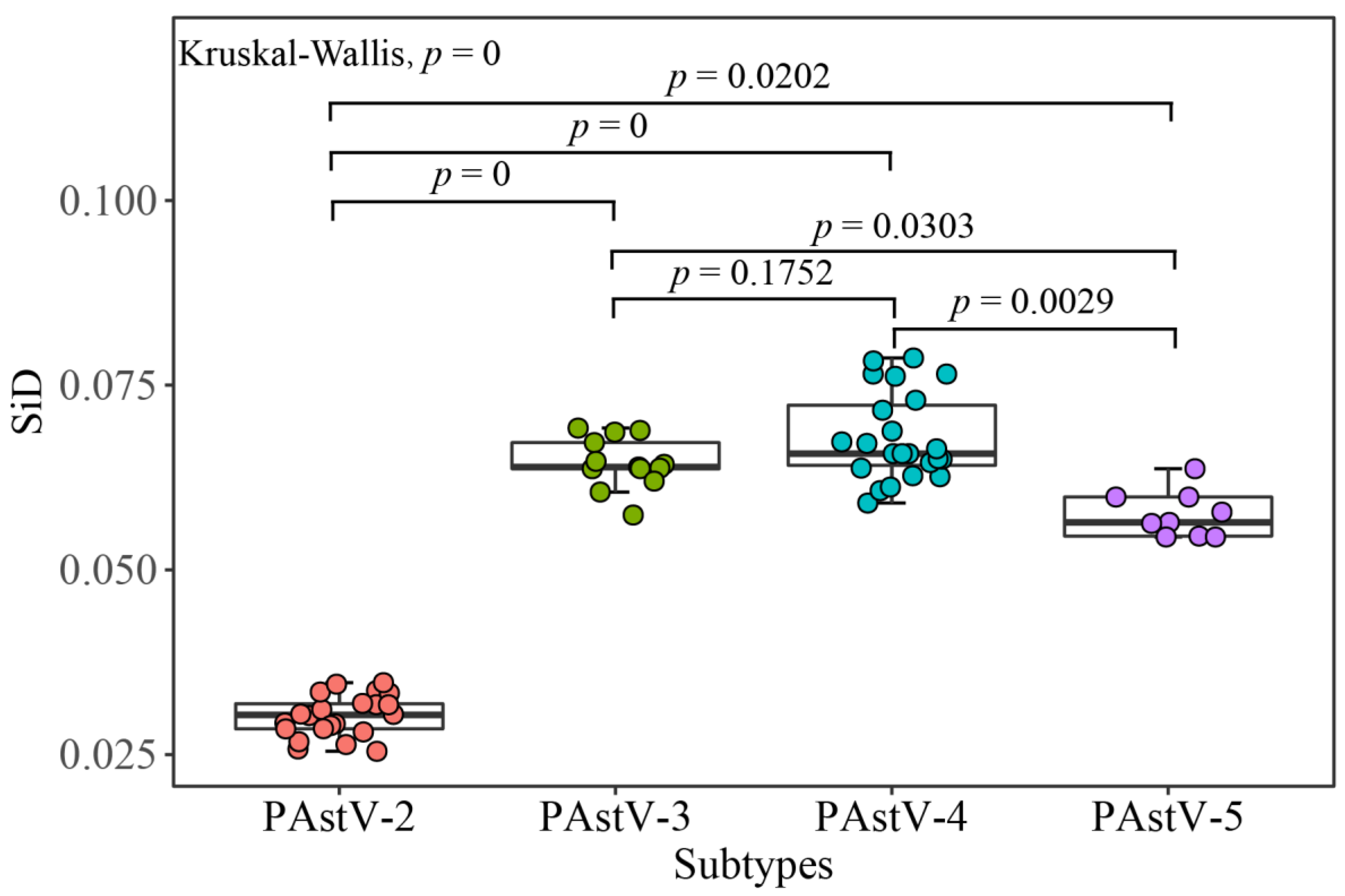
2.12. Similarity Index (SiD) Analysis
2.13. Statistical Analysis
3. Results
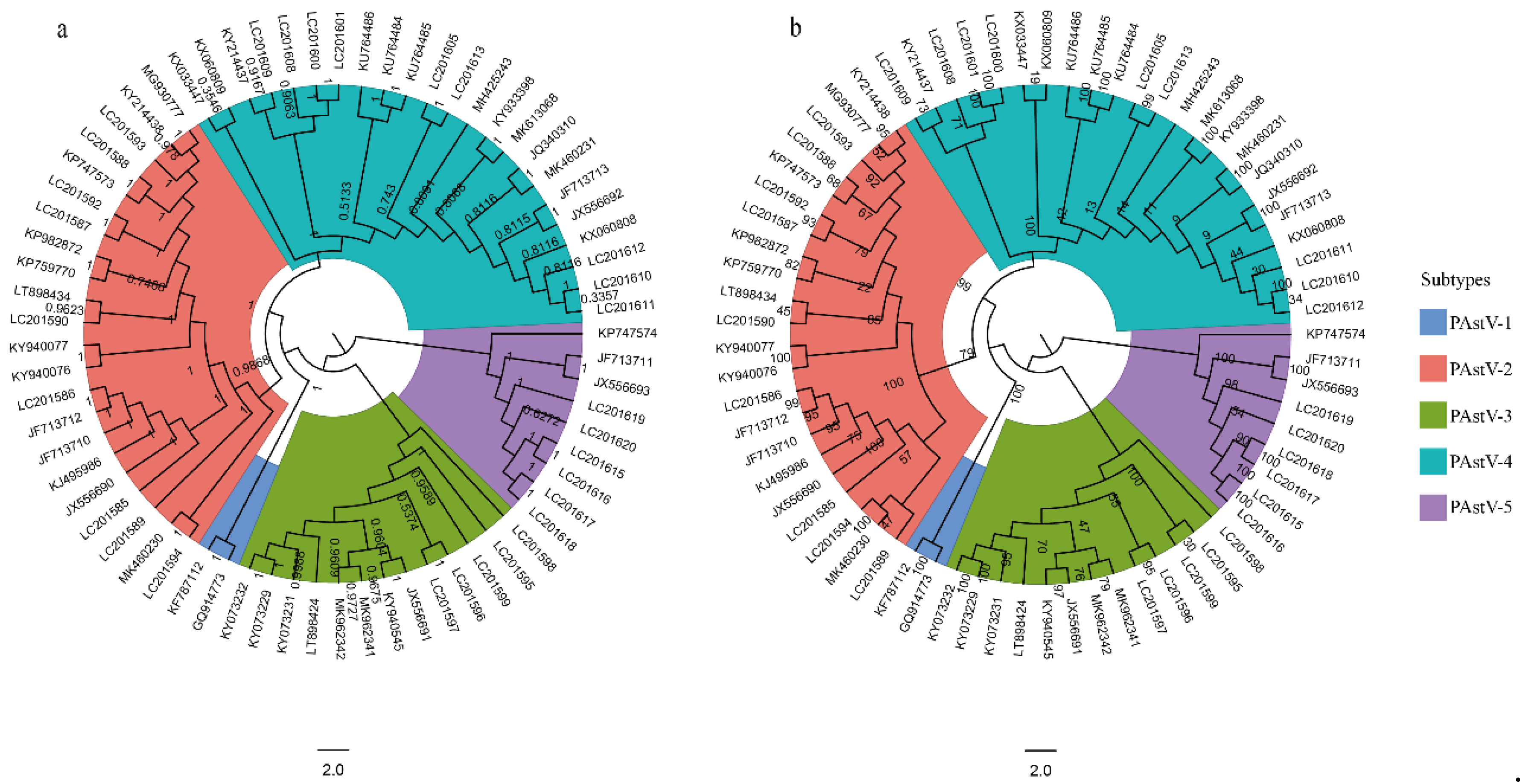
3.1. PAstV-5 Subtype Has the Basal Position of Phylogenetic Tree
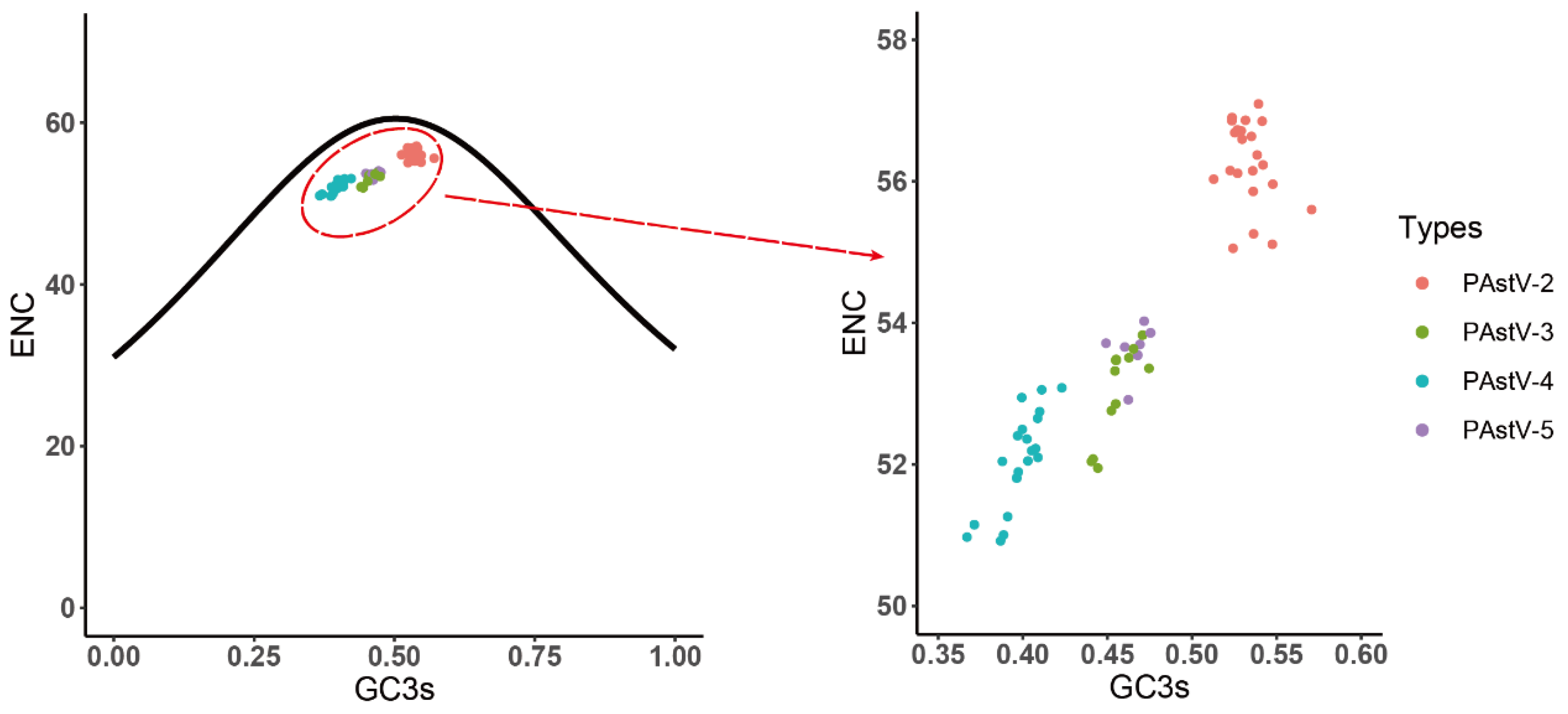
3.2. A and U Nucleotides Were More Frequent than C and G in the PAstV Coding Sequences
3.3. RSCU Patterns of PAstV
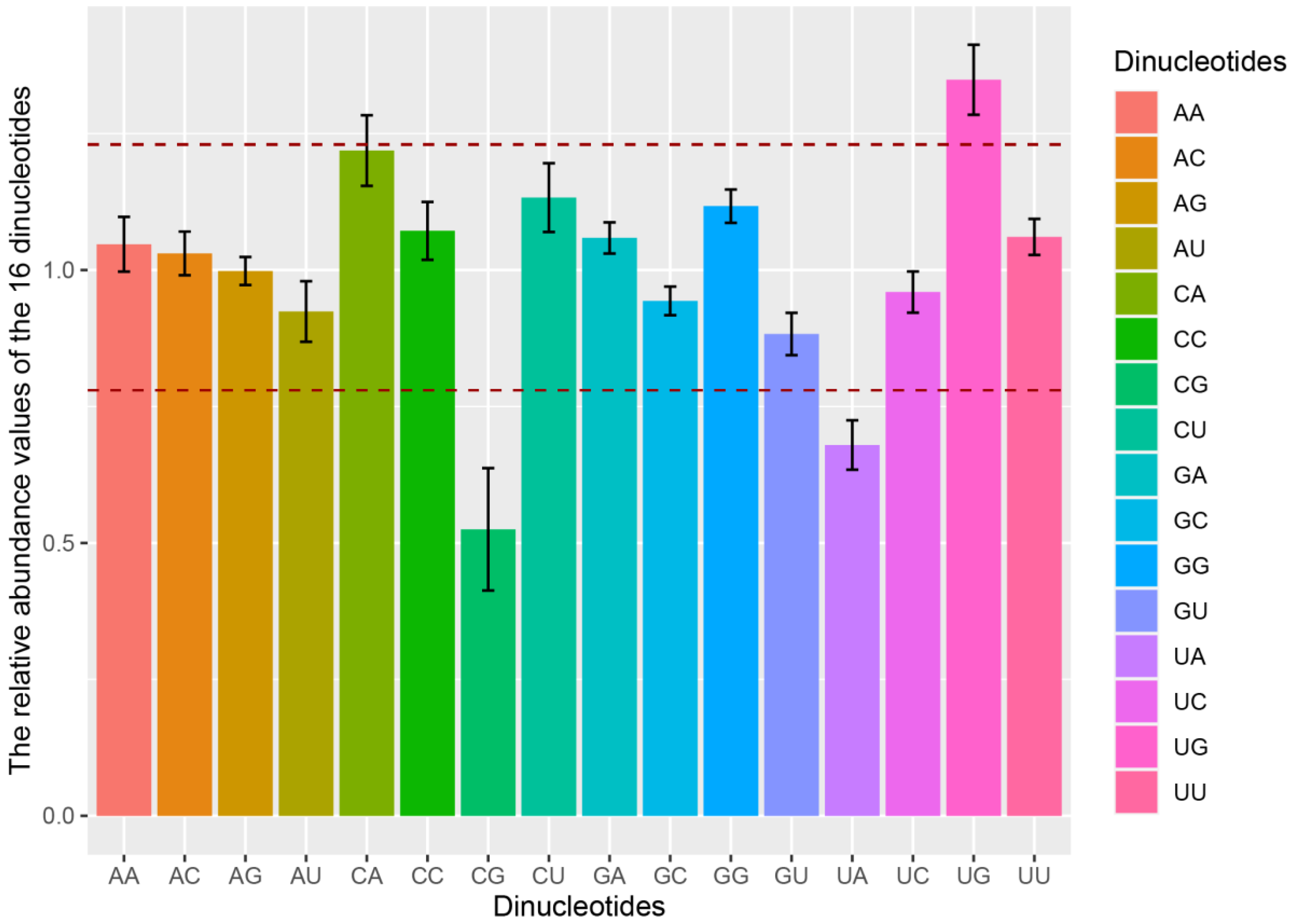
3.4. Dinucleotide Frequency Abundancy Influences the Codon Usage Bias of PAstV
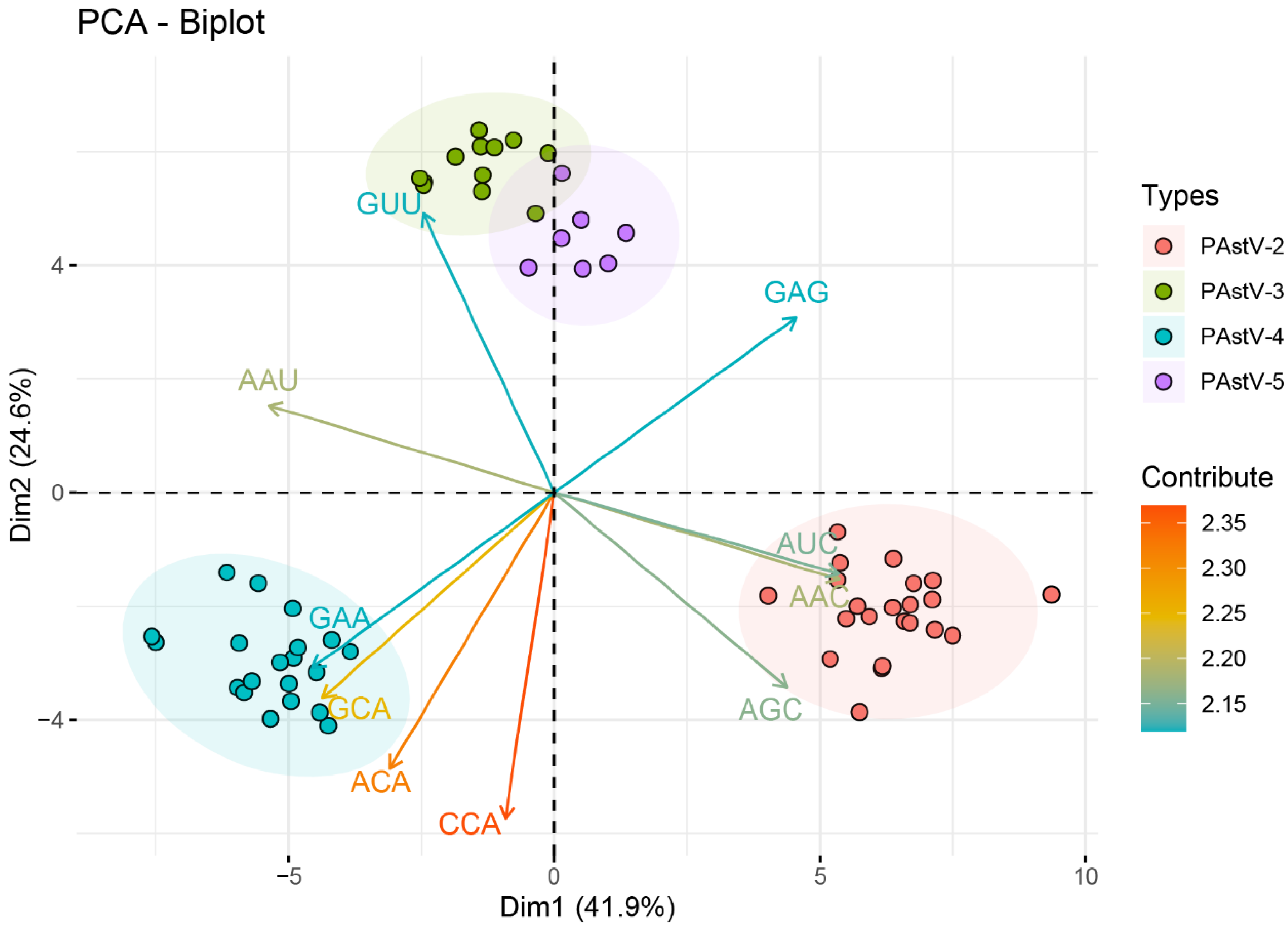
3.5. Identification of the Forces Influencing Codon Usage Patterns
3.6. PAstV Strains Adaptation to Host Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Reuter, G.; Knowles, N.J. Astroviruses. Dis. Swine 2019, 457–460. [Google Scholar] [CrossRef]
- Bridger, J. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet. Rec. 1980, 107, 532–533. [Google Scholar] [PubMed]
- Luo, Z.; Roi, S.; Dastor, M.; Gallice, E.; Laurin, M.-A.; L’Homme, Y. Multiple novel and prevalent astroviruses in pigs. Vet. Microbiol. 2011, 149, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Bailey Arruda, P.A.; Hensch, M.; Chen, Q.; Zheng, Y.; Yang, C.; Gatto, I.R.H.; Ferreyra, F.M.; Gauger, P.; Schwartz, K.; Bradner, L. Porcine astrovirus type 3 in central nervous system of swine with polioencephalomyelitis. Emerg. Infect. Dis. 2017, 23, 2097. [Google Scholar] [CrossRef] [Green Version]
- Boros, Á.; Albert, M.; Pankovics, P.; Bíró, H.; Pesavento, P.A.; Phan, T.G.; Delwart, E.; Reuter, G. Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, Hungary. Emerg. Infect. Dis. 2017, 23, 1982. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, C.; Liu, H.; Wu, Q.; Liang, S.; Cen, M.; Dong, Q.; Wei, Y.; Chen, Y.; Ouyang, K. Pathogenic Characteristics of a Porcine Astrovirus Strain Isolated in China. Viruses 2019, 11, 1156. [Google Scholar] [CrossRef] [Green Version]
- Padmanabhan, A.; Hause, B.M. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Arch. Virol. 2016, 161, 2575–2579. [Google Scholar] [CrossRef]
- Kumthip, K.; Khamrin, P.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Ushijima, H.; Maneekarn, N. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch. Virol. 2018, 163, 1823–1829. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Gimenez-Lirola, L.G.; Gerber, P.F.; Jiang, Y.-H.; Halbur, P.G.; Opriessnig, T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J. Gen. Virol. 2013, 94, 570–582. [Google Scholar] [CrossRef]
- Blomström, A.-L.; Ley, C.; Jacobson, M. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta Vet. Scand. 2014, 56, 82. [Google Scholar] [CrossRef] [Green Version]
- Brnić, D.; Prpić, J.; Keros, T.; Roić, B.; Starešina, V.; Jemeršić, L. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect. Genet. Evol. 2013, 14, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Monroe, S.S.; Koonin, E.V.; Stine, S.E.; Glass, R.I. RNA sequence of astrovirus: Distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 1993, 90, 10539–10543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marczinke, B.; Bloys, A.J.; Brown, T.; Willcocks, M.M.; Carter, M.J.; Brierley, I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 1994, 68, 5588–5595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arias, C.F.; DuBois, R.M. The astrovirus capsid: A review. Viruses 2017, 9, 15. [Google Scholar] [CrossRef]
- Sharp, P.M.; Matassi, G. Codon usage and genome evolution. Curr. Opin. Genet. Dev. 1994, 4, 851–860. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef]
- Van Hemert, F.J.; Berkhout, B.; Lukashov, V.V. Host-related nucleotide composition and codon usage as driving forces in the recent evolution of the Astroviridae. Virology 2007, 361, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Ranwez, V.; Harispe, S.; Delsuc, F.; Douzery, E.J. MACSE: Multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS ONE 2011, 6, e22594. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A. 2012. FigTree v1. 4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 5 September 2020).
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 5 September 2020).
- Kariin, S.; Burge, C. Dinucleotide relative abundance extremes: A genomic signature. Trends Genet. 1995, 11, 283–290. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Fuglsang, A. The ‘effective number of codons’ revisited. Biochem. Biophys. Res. Commun. 2004, 317, 957–964. [Google Scholar] [CrossRef]
- Elek, A.; Kuzman, M.; Vlahoviček, K. coRdon: Codon Usage Analysis and Prediction of Gene Expressivity. R Package Version 1.4.0. 2019. Available online: https://github.com/BioinfoHR/coRdon (accessed on 5 September 2020).
- Wong, E.H.; Smith, D.K.; Rabadan, R.; Peiris, M.; Poon, L.L. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. BMC Evol. Biol. 2010, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Sharp, P.M.; Tuohy, T.M.; Mosurski, K.R. Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986, 14, 5125–5143. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. R Package Version 2017, 1, 2017. [Google Scholar]
- Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef] [Green Version]
- Sueoka, N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G+ C content of third codon position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Sueoka, N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct 2008, 3, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santana-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A new and updated resource for codon usage tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, S.; Papamichail, D.; Coleman, J.R.; Skiena, S.; Wimmer, E. Reduction of the Rate of Poliovirus Protein Synthesis through Large-Scale Codon Deoptimization Causes Attenuation of Viral Virulence by Lowering Specific Infectivity. J. Virol. 2006, 80, 9687–9696. [Google Scholar] [CrossRef] [Green Version]
- Puigbò, P.; Aragonès, L.; Garcia-Vallvé, S. RCDI/eRCDI: A web-server to estimate codon usage deoptimization. BMC Res. Notes 2010, 3, 87. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, J.; Sun, D.; Ma, Q.; Chen, H.; Ma, L.; Ding, Y.; Liu, Y. The distribution of synonymous codon choice in the translation initiation region of dengue virus. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Dinno, A. Dunn.Test: Dunn’s Test of Multiple Comparisons Using Rank Sums. R Package Version 1.3.5. 2017. Available online: https://CRAN.R-project.org/package=dunn.test (accessed on 5 September 2020).
- Streicker, D.G. From persistence to cross-species emergence of a viral zoonosis. Science 2013, 342, 1185–1186. [Google Scholar] [CrossRef] [Green Version]
- Salim, H.M.; Cavalcanti, A.R. Factors influencing codon usage bias in genomes. J. Braz. Chem. Soc. 2008, 19, 257–262. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wang, H.; Wang, S.; Xing, G.; Zhang, C.; Zhang, W.; Liu, J.; Zhang, J.; Su, S.; Zhou, J. Insights into the genetic and host adaptability of emerging porcine circovirus 3. Virulence 2018, 9, 1301–1313. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Wang, N.; Tan, J.; Wang, R.; Yang, Y.; Li, G.; Guan, H.; Zheng, Y.; Shi, X.; Ye, R. Comprehensive codon usage analysis of porcine deltacoronavirus. Mol. Phylogenet. Evol. 2019, 141, 106618. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, Y.; Zhai, X.; Liu, J.; Zhao, W.; Ji, S.; Su, S.; Zhou, J. Comprehensive analysis of codon usage on rabies virus and other lyssaviruses. Int. J. Mol. Sci. 2018, 19, 2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, A.M.; Nasrullah, I.; Qamar, R.; Tong, Y. Evolution of codon usage in Zika virus genomes is host and vector specific. Emerg. Microbes Infect. 2016, 5, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Bera, B.C.; Greenbaum, B.D.; Bhatia, S.; Sood, R.; Selvaraj, P.; Anand, T.; Tripathi, B.N.; Virmani, N. Revelation of influencing factors in overall codon usage bias of equine influenza viruses. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, A.M.; Nasrullah, I.; Tong, Y. Genome-wide analysis of codon usage and influencing factors in chikungunya viruses. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [Green Version]
- Khandia, R.; Singhal, S.; Kumar, U.; Ansari, A.I.; Tiwari, R.; Dhama, K.; Das, J.; Munjal, A.; Singh, R.K. Analysis of Nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019, 10, 886. [Google Scholar] [CrossRef] [Green Version]
- Yomo, T.; Ohno, S. Concordant evolution of coding and noncoding regions of DNA made possible by the universal rule of TA/CG deficiency-TG/CT excess. Proc. Natl. Acad. Sci. USA 1989, 86, 8452–8456. [Google Scholar] [CrossRef] [Green Version]
- Rima, B.K.; McFerran, N.V. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol. 1997, 78, 2859–2870. [Google Scholar] [CrossRef]
- Vetsigian, K.; Goldenfeld, N. Genome rhetoric and the emergence of compositional bias. Proc. Natl. Acad. Sci. USA 2009, 106, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Breslauer, K.J.; Frank, R.; Blöcker, H.; Marky, L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Dong, H.; Jiang, C.; Cao, F.; Tao, S.; Xu, L. Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Sci. Rep. 2016, 6, 35927. [Google Scholar] [CrossRef]
- Jia, X.; Liu, S.; Zheng, H.; Li, B.; Qi, Q.; Wei, L.; Zhao, T.; He, J.; Sun, J. Non-uniqueness of factors constraint on the codon usage in Bombyx mori. BMC Genom. 2015, 16, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawabe, A.; Miyashita, N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003, 78, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, C.; Vijaykrishna, D. The broad host range and genetic diversity of mammalian and avian astroviruses. Viruses 2017, 9, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wohlgemuth, N.; Honce, R.; Schultz-Cherry, S. Astrovirus evolution and emergence. Infect. Genet. Evol. 2019, 69, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kuroda, M.; Masuda, T.; Akagami, M.; Haga, K.; Tsuchiaka, S.; Kishimoto, M.; Naoi, Y.; Sano, K.; Omatsu, T. Whole genome analysis of porcine astroviruses detected in Japanese pigs reveals genetic diversity and possible intra-genotypic recombination. Infect. Genet. Evol. 2017, 50, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Kunec, D.; Osterrieder, N. Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep. 2016, 14, 55–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef]
- Coleman, J.R.; Papamichail, D.; Skiena, S.; Futcher, B.; Wimmer, E.; Mueller, S. Virus attenuation by genome-scale changes in codon pair bias. Science 2008, 320, 1784–1787. [Google Scholar] [CrossRef] [Green Version]
- Burns, C.C.; Shaw, J.; Campagnoli, R.; Jorba, J.; Vincent, A.; Quay, J.; Kew, O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 2006, 80, 3259–3272. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.C.; Bischof, G.F.; Lauer, W.A.; Desrosiers, R.C. Importance of codon usage for the temporal regulation of viral gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 14030–14035. [Google Scholar] [CrossRef] [Green Version]


| Catalogs | PAstV-2 | PAstV-3 | PAstV-4 | PAstV-5 | All |
|---|---|---|---|---|---|
| A | 0.268 ± 0.004 | 0.247 ± 0.002 | 0.301 ± 0.003 | 0.238 ± 0.002 | 0.271 ± 0.024 |
| C | 0.263 ± 0.004 | 0.234 ± 0.003 | 0.222 ± 0.004 | 0.259 ± 0.005 | 0.243 ± 0.019 |
| G | 0.244 ± 0.003 | 0.24 ± 0.002 | 0.227 ± 0.003 | 0.234 ± 0.001 | 0.236 ± 0.008 |
| U | 0.225 ± 0.004 | 0.279 ± 0.002 | 0.25 ± 0.004 | 0.269 ± 0.004 | 0.25 ± 0.021 |
| GC | 0.507 ± 0.005 | 0.474 ± 0.004 | 0.449 ± 0.006 | 0.493 ± 0.005 | 0.479 ± 0.025 |
| GC1s | 0.549 ± 0.006 | 0.541 ± 0.003 | 0.528 ± 0.004 | 0.565 ± 0.007 | 0.543 ± 0.014 |
| GC2s | 0.438 ± 0.004 | 0.424 ± 0.003 | 0.422 ± 0.007 | 0.448 ± 0.003 | 0.431 ± 0.011 |
| GC12s | 0.494 ± 0.003 | 0.483 ± 0.003 | 0.475 ± 0.005 | 0.506 ± 0.004 | 0.487 ± 0.012 |
| GC3s | 0.534 ± 0.012 | 0.456 ± 0.01 | 0.397 ± 0.014 | 0.467 ± 0.008 | 0.463 ± 0.058 |
| U3s | 0.308 ± 0.014 | 0.433 ± 0.01 | 0.399 ± 0.011 | 0.421 ± 0.009 | 0.379 ± 0.052 |
| C3s | 0.371 ± 0.011 | 0.283 ± 0.008 | 0.241 ± 0.013 | 0.312 ± 0.011 | 0.302 ± 0.056 |
| A3s | 0.286 ± 0.011 | 0.256 ± 0.006 | 0.384 ± 0.01 | 0.245 ± 0.006 | 0.308 ± 0.057 |
| G3s | 0.276 ± 0.009 | 0.255 ± 0.008 | 0.235 ± 0.008 | 0.237 ± 0.005 | 0.253 ± 0.02 |
| ENC | 56.265 ± 0.602 | 53.059 ± 0.656 | 52.007 ± 0.678 | 53.647 ± 0.316 | 53.83 ± 1.902 |
| Codon | PAstV-2 | PAstV-3 | PAstV-4 | PAstV-5 | All(G) |
|---|---|---|---|---|---|
| GCA(A) | 1.129 ± 0.124 | 1.097 ± 0.084 | 1.813 ± 0.159 | 1.016 ± 0.05 | 1.343 ± 0.366 |
| GCC(A) | 1.331 ± 0.131 | 1.202 ± 0.074 | 0.874 ± 0.099 | 1.266 ± 0.103 | 1.14 ± 0.225 |
| GCG(A) | 0.568 ± 0.064 | 0.365 ± 0.061 | 0.34 ± 0.058 | 0.275 ± 0.049 | 0.411 ± 0.128 |
| GCU(A) | 0.972 ± 0.15 | 1.335 ± 0.09 | 0.972 ± 0.124 | 1.443 ± 0.082 | 1.106 ± 0.23 |
| UGC(C) | 0.98 ± 0.171 | 0.781 ± 0.187 | 0.71 ± 0.1 | 0.895 ± 0.18 | 0.837 ± 0.191 |
| UGU(C) | 1.02 ± 0.171 | 1.219 ± 0.187 | 1.29 ± 0.1 | 1.105 ± 0.18 | 1.163 ± 0.191 |
| GAC(D) | 0.961 ± 0.086 | 0.803 ± 0.06 | 0.692 ± 0.077 | 0.979 ± 0.102 | 0.84 ± 0.147 |
| GAU(D) | 1.039 ± 0.086 | 1.197 ± 0.06 | 1.308 ± 0.077 | 1.021 ± 0.102 | 1.16 ± 0.147 |
| GAA(E) | 0.834 ± 0.08 | 0.825 ± 0.08 | 1.163 ± 0.077 | 0.884 ± 0.075 | 0.952 ± 0.172 |
| GAG(E) | 1.166 ± 0.08 | 1.175 ± 0.08 | 0.837 ± 0.077 | 1.116 ± 0.075 | 1.048 ± 0.172 |
| UUC(F) | 1.072 ± 0.096 | 0.679 ± 0.055 | 0.702 ± 0.099 | 0.874 ± 0.121 | 0.842 ± 0.196 |
| UUU(F) | 0.928 ± 0.096 | 1.321 ± 0.055 | 1.298 ± 0.099 | 1.126 ± 0.121 | 1.158 ± 0.196 |
| GGA(G) | 0.791 ± 0.118 | 0.531 ± 0.081 | 1.055 ± 0.092 | 0.486 ± 0.086 | 0.79 ± 0.244 |
| GGC(G) | 1.189 ± 0.117 | 1.332 ± 0.119 | 0.896 ± 0.119 | 1.092 ± 0.065 | 1.103 ± 0.2 |
| GGG(G) | 0.88 ± 0.138 | 0.629 ± 0.065 | 0.729 ± 0.079 | 0.624 ± 0.099 | 0.745 ± 0.144 |
| GGU(G) | 1.14 ± 0.146 | 1.508 ± 0.069 | 1.32 ± 0.124 | 1.798 ± 0.1 | 1.362 ± 0.247 |
| CAC(H) | 1.183 ± 0.127 | 0.898 ± 0.105 | 0.688 ± 0.087 | 0.775 ± 0.068 | 0.903 ± 0.234 |
| CAU(H) | 0.817 ± 0.127 | 1.102 ± 0.105 | 1.312 ± 0.087 | 1.225 ± 0.068 | 1.097 ± 0.234 |
| AUA(I) | 0.738 ± 0.109 | 0.652 ± 0.1 | 0.783 ± 0.11 | 0.889 ± 0.028 | 0.757 ± 0.121 |
| AUC(I) | 1.356 ± 0.107 | 0.93 ± 0.048 | 0.916 ± 0.132 | 1.07 ± 0.034 | 1.084 ± 0.222 |
| AUU(I) | 0.906 ± 0.116 | 1.418 ± 0.097 | 1.301 ± 0.105 | 1.041 ± 0.033 | 1.159 ± 0.231 |
| AAA(K) | 0.927 ± 0.093 | 0.86 ± 0.037 | 0.989 ± 0.08 | 0.954 ± 0.036 | 0.939 ± 0.087 |
| AAG(K) | 1.073 ± 0.093 | 1.14 ± 0.037 | 1.011 ± 0.08 | 1.046 ± 0.036 | 1.061 ± 0.087 |
| CUA(L) | 0.635 ± 0.163 | 0.501 ± 0.079 | 0.561 ± 0.128 | 0.425 ± 0.067 | 0.555 ± 0.144 |
| CUC(L) | 1.639 ± 0.161 | 0.849 ± 0.062 | 1.127 ± 0.141 | 1.361 ± 0.132 | 1.273 ± 0.326 |
| CUG(L) | 0.953 ± 0.17 | 0.668 ± 0.062 | 0.699 ± 0.083 | 0.83 ± 0.089 | 0.794 ± 0.168 |
| CUU(L) | 1.582 ± 0.155 | 1.915 ± 0.093 | 1.886 ± 0.156 | 1.814 ± 0.215 | 1.782 ± 0.209 |
| UUA(L) | 0.404 ± 0.093 | 0.889 ± 0.127 | 0.715 ± 0.09 | 0.458 ± 0.121 | 0.612 ± 0.216 |
| UUG(L) | 0.787 ± 0.141 | 1.178 ± 0.138 | 1.011 ± 0.137 | 1.112 ± 0.105 | 0.983 ± 0.2 |
| AAC(N) | 1.213 ± 0.107 | 0.819 ± 0.09 | 0.774 ± 0.102 | 0.868 ± 0.063 | 0.939 ± 0.217 |
| AAU(N) | 0.787 ± 0.107 | 1.181 ± 0.09 | 1.226 ± 0.102 | 1.132 ± 0.063 | 1.061 ± 0.217 |
| CCA(P) | 1.802 ± 0.115 | 1.084 ± 0.07 | 2.029 ± 0.111 | 1.271 ± 0.066 | 1.669 ± 0.387 |
| CCC(P) | 0.789 ± 0.102 | 0.876 ± 0.049 | 0.49 ± 0.112 | 0.903 ± 0.077 | 0.718 ± 0.195 |
| CCG(P) | 0.393 ± 0.099 | 0.308 ± 0.038 | 0.218 ± 0.053 | 0.296 ± 0.052 | 0.303 ± 0.1 |
| CCU(P) | 1.016 ± 0.114 | 1.733 ± 0.082 | 1.264 ± 0.139 | 1.531 ± 0.094 | 1.309 ± 0.29 |
| CAA(Q) | 0.836 ± 0.118 | 1.068 ± 0.087 | 1.048 ± 0.075 | 0.899 ± 0.098 | 0.962 ± 0.14 |
| CAG(Q) | 1.164 ± 0.118 | 0.932 ± 0.087 | 0.952 ± 0.075 | 1.101 ± 0.098 | 1.038 ± 0.14 |
| AGA(R) | 0.815 ± 0.183 | 0.555 ± 0.083 | 1.458 ± 0.166 | 0.726 ± 0.127 | 0.973 ± 0.395 |
| AGG(R) | 1.298 ± 0.119 | 1.43 ± 0.121 | 1.865 ± 0.209 | 1.183 ± 0.061 | 1.503 ± 0.311 |
| CGA(R) | 0.479 ± 0.119 | 0.375 ± 0.079 | 0.401 ± 0.118 | 0.354 ± 0.047 | 0.415 ± 0.113 |
| CGC(R) | 1.486 ± 0.225 | 1.292 ± 0.14 | 0.803 ± 0.136 | 1.478 ± 0.152 | 1.213 ± 0.35 |
| CGG(R) | 0.852 ± 0.156 | 0.609 ± 0.078 | 0.425 ± 0.091 | 0.585 ± 0.093 | 0.622 ± 0.21 |
| CGU(R) | 1.07 ± 0.174 | 1.739 ± 0.142 | 1.048 ± 0.188 | 1.674 ± 0.169 | 1.274 ± 0.353 |
| AGC(S) | 1.005 ± 0.136 | 0.222 ± 0.044 | 0.465 ± 0.125 | 0.386 ± 0.058 | 0.584 ± 0.327 |
| AGU(S) | 0.819 ± 0.16 | 0.313 ± 0.069 | 1.008 ± 0.11 | 0.651 ± 0.105 | 0.763 ± 0.279 |
| UCA(S) | 1.442 ± 0.227 | 1.549 ± 0.147 | 2.068 ± 0.238 | 1.5 ± 0.097 | 1.685 ± 0.345 |
| UCC(S) | 1.184 ± 0.2 | 1.404 ± 0.181 | 0.755 ± 0.158 | 1.157 ± 0.098 | 1.076 ± 0.3 |
| UCG(S) | 0.48 ± 0.145 | 0.511 ± 0.14 | 0.225 ± 0.065 | 0.434 ± 0.099 | 0.392 ± 0.168 |
| UCU(S) | 1.069 ± 0.194 | 2.001 ± 0.171 | 1.48 ± 0.231 | 1.873 ± 0.131 | 1.499 ± 0.409 |
| ACA(U) | 1.347 ± 0.194 | 0.995 ± 0.091 | 2.023 ± 0.133 | 0.854 ± 0.048 | 1.445 ± 0.476 |
| ACC(U) | 1.314 ± 0.157 | 1.099 ± 0.096 | 0.717 ± 0.094 | 1.266 ± 0.095 | 1.061 ± 0.287 |
| ACG(U) | 0.39 ± 0.099 | 0.348 ± 0.087 | 0.182 ± 0.053 | 0.279 ± 0.066 | 0.296 ± 0.118 |
| ACU(U) | 0.949 ± 0.143 | 1.559 ± 0.096 | 1.078 ± 0.106 | 1.601 ± 0.111 | 1.199 ± 0.295 |
| GUA(V) | 0.418 ± 0.113 | 0.406 ± 0.044 | 0.713 ± 0.141 | 0.241 ± 0.038 | 0.493 ± 0.2 |
| GUC(V) | 1.309 ± 0.162 | 0.894 ± 0.087 | 0.918 ± 0.139 | 1.079 ± 0.117 | 1.064 ± 0.226 |
| GUG(V) | 1.164 ± 0.117 | 0.936 ± 0.103 | 1.004 ± 0.133 | 0.834 ± 0.117 | 1.021 ± 0.164 |
| GUU(V) | 1.109 ± 0.16 | 1.764 ± 0.072 | 1.365 ± 0.144 | 1.847 ± 0.159 | 1.423 ± 0.317 |
| UAC(Y) | 1.238 ± 0.092 | 0.815 ± 0.127 | 0.775 ± 0.11 | 0.812 ± 0.038 | 0.94 ± 0.233 |
| UAU(Y) | 0.762 ± 0.092 | 1.185 ± 0.127 | 1.225 ± 0.11 | 1.188 ± 0.038 | 1.06 ± 0.233 |
| PAstV-2 | PAstV-3 | PAstV-4 | PAstV-5 | All(G) | |
|---|---|---|---|---|---|
| AA | 1.001 ± 0.033 | 1.121 ± 0.015 | 1.039 ± 0.021 | 1.076 ± 0.021 | 1.047 ± 0.05 |
| AC | 1.049 ± 0.022 | 1.025 ± 0.021 | 0.991 ± 0.02 | 1.092 ± 0.017 | 1.03 ± 0.04 |
| AG | 0.977 ± 0.025 | 1 ± 0.018 | 1.011 ± 0.02 | 1.014 ± 0.014 | 0.998 ± 0.026 |
| AU | 0.967 ± 0.025 | 0.872 ± 0.021 | 0.949 ± 0.019 | 0.83 ± 0.026 | 0.924 ± 0.055 |
| CA | 1.239 ± 0.023 | 1.137 ± 0.027 | 1.279 ± 0.022 | 1.135 ± 0.022 | 1.219 ± 0.065 |
| CC | 1.028 ± 0.022 | 1.124 ± 0.028 | 1.11 ± 0.021 | 1.003 ± 0.021 | 1.072 ± 0.053 |
| CG | 0.642 ± 0.027 | 0.528 ± 0.023 | 0.384 ± 0.015 | 0.595 ± 0.01 | 0.525 ± 0.112 |
| CU | 1.072 ± 0.03 | 1.181 ± 0.02 | 1.126 ± 0.041 | 1.23 ± 0.02 | 1.133 ± 0.063 |
| GA | 1.055 ± 0.024 | 1.032 ± 0.023 | 1.062 ± 0.021 | 1.1 ± 0.011 | 1.059 ± 0.029 |
| GC | 0.928 ± 0.021 | 0.955 ± 0.023 | 0.954 ± 0.029 | 0.937 ± 0.017 | 0.943 ± 0.026 |
| GG | 1.09 ± 0.026 | 1.142 ± 0.009 | 1.129 ± 0.025 | 1.118 ± 0.026 | 1.117 ± 0.031 |
| GU | 0.92 ± 0.03 | 0.886 ± 0.022 | 0.851 ± 0.029 | 0.871 ± 0.016 | 0.883 ± 0.039 |
| UA | 0.659 ± 0.021 | 0.748 ± 0.023 | 0.647 ± 0.022 | 0.713 ± 0.02 | 0.679 ± 0.045 |
| UC | 0.987 ± 0.032 | 0.913 ± 0.014 | 0.956 ± 0.028 | 0.971 ± 0.027 | 0.96 ± 0.038 |
| UG | 1.349 ± 0.031 | 1.274 ± 0.02 | 1.418 ± 0.025 | 1.277 ± 0.018 | 1.349 ± 0.064 |
| UU | 1.043 ± 0.028 | 1.061 ± 0.019 | 1.085 ± 0.031 | 1.041 ± 0.024 | 1.061 ± 0.033 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Bao, Z.; Mou, C.; Chen, Z.; Zhao, J. Comprehensive Analysis of Codon Usage on Porcine Astrovirus. Viruses 2020, 12, 991. https://doi.org/10.3390/v12090991
Wu H, Bao Z, Mou C, Chen Z, Zhao J. Comprehensive Analysis of Codon Usage on Porcine Astrovirus. Viruses. 2020; 12(9):991. https://doi.org/10.3390/v12090991
Chicago/Turabian StyleWu, Huiguang, Zhengyu Bao, Chunxiao Mou, Zhenhai Chen, and Jingwen Zhao. 2020. "Comprehensive Analysis of Codon Usage on Porcine Astrovirus" Viruses 12, no. 9: 991. https://doi.org/10.3390/v12090991




