The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies
Abstract
:1. Introduction
2. Capsid Assembly
2.1. Overview of Capsid Components and Experimental Assembly Systems
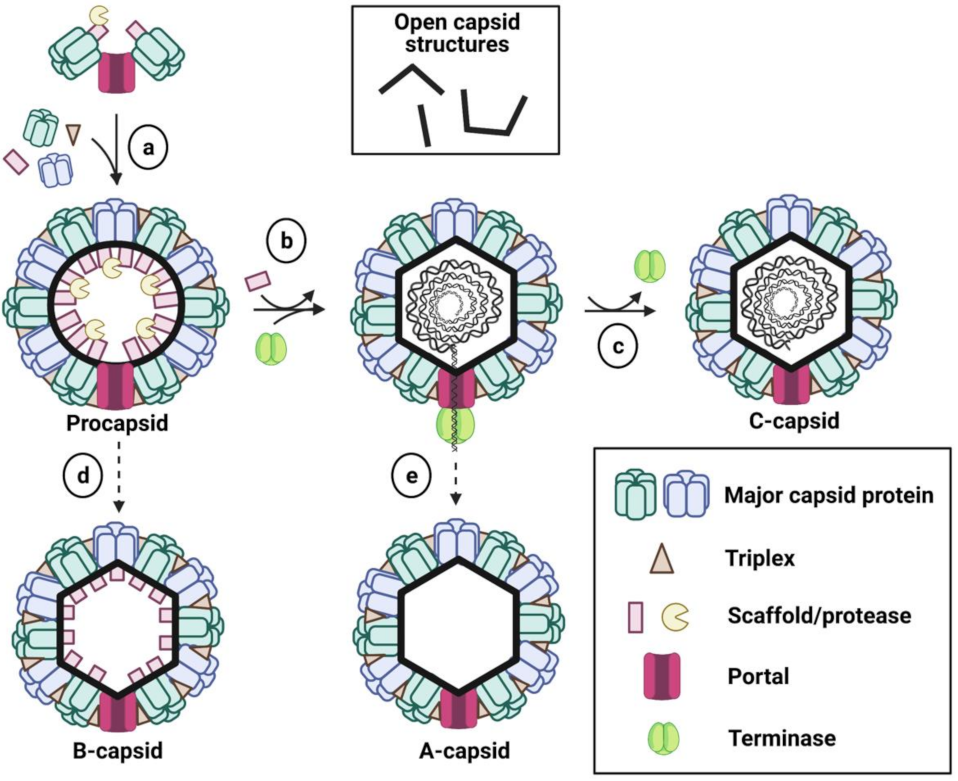
2.2. Capsid Assembly Pathway
2.3. Sub-Family-Specific Roles of Capsid Proteins in Alphaherpesvirus Assembly
2.3.1. MCP, Triplexes, the Formation of Procapsids, and the Portal
| Alpha | Beta | Gamma | ||||
|---|---|---|---|---|---|---|
| HSV-1 | HCMV | EBV/KSHV | ||||
| ∆Protein | In Vitro | Insect Cells | Infected Cells | Infected Cells | Insect Cells | Infected Cells |
| ∆MCP | No capsid formation [23] | No capsid formation [19,20] | No capsid formation [40] | No viral replication (capsid assembly not investigated) [42] | No capsid formation [21,22] | -- |
| ∆SCP | B-capsids [23] | B-capsids [19,20] | A-, B- and C-capsids form in Vero cells and ocular mouse model b [43] | B-capsids [44] | No capsid formation [21,22] | Severe defect in capsid formation; some empty capsids (KSHV) [45,46] |
| ∆Triplex 1 | No capsid formation [23] | No capsid formation [19,20] | No capsid formation [41] | No viral replication (capsid assembly not investigated) [42] | No capsid formation [21,22] | -- |
| ∆Triplex 2 | No capsid formation [23] | No capsid formation [19,20] | No capsid formation [40] | No viral replication (capsid assembly not investigated) [42] | No capsid formation [21,22] | -- |
| ∆Scaffold | -- | Empty angularized capsids and open shells [19,20] | B-capsids [47] | Some closed capsids of unknown identity; open capsid shells [48] | No capsid formation; only open capsid structures [21,22] | -- |
| ∆Protease | Capsids form at a reduced yield; capsid type was not determined a [23] | Angularized capsids, similar in appearance to B-capsids [19,20] | Only procapsids; these are capable of maturation if isolated and incubated at room temperature [30,49] | Angularized capsids, similar in appearance to B-capsids with dense cores [50] | B-capsids [21,22] | Only closed spherical procapsids (KSHV) [51] |
| ∆Scaffold + ∆Protease | No capsid formation [23] | No capsid formation; only open capsid structures [19,20] | No capsid formation; only open capsid structures [52] | -- | No capsid formation; only open capsid structures [21,22] | -- |
| ∆Portal | B-capsids [23] | B-capsids [19,20] | B-capsids either when deleted [53] or portal/scaffold interactions perturbed [54,55] | -- | B-capsids [21,22] | -- |
2.3.2. Protease and Scaffold
2.3.3. SCP
2.4. Sub-Family-Specific Roles of Capsid Proteins in Gammaherpesvirus Assembly
2.4.1. MCP and Triplexes
2.4.2. Protease and Scaffold
2.4.3. SCP
2.5. Sub-Family-Specific Roles of Capsid Proteins in Betaherpesvirus Assembly
2.5.1. MCP and Triplexes
2.5.2. Protease and Scaffold
2.5.3. SCP
2.6. Summary of Capsid Assembly across Subfamilies
3. Capsids Require Additional Capsid-Associated Proteins for Successful Viral Replication
3.1. Alphaherpesviruses
3.1.1. CATC Components, Capsid Location, and Occupancy
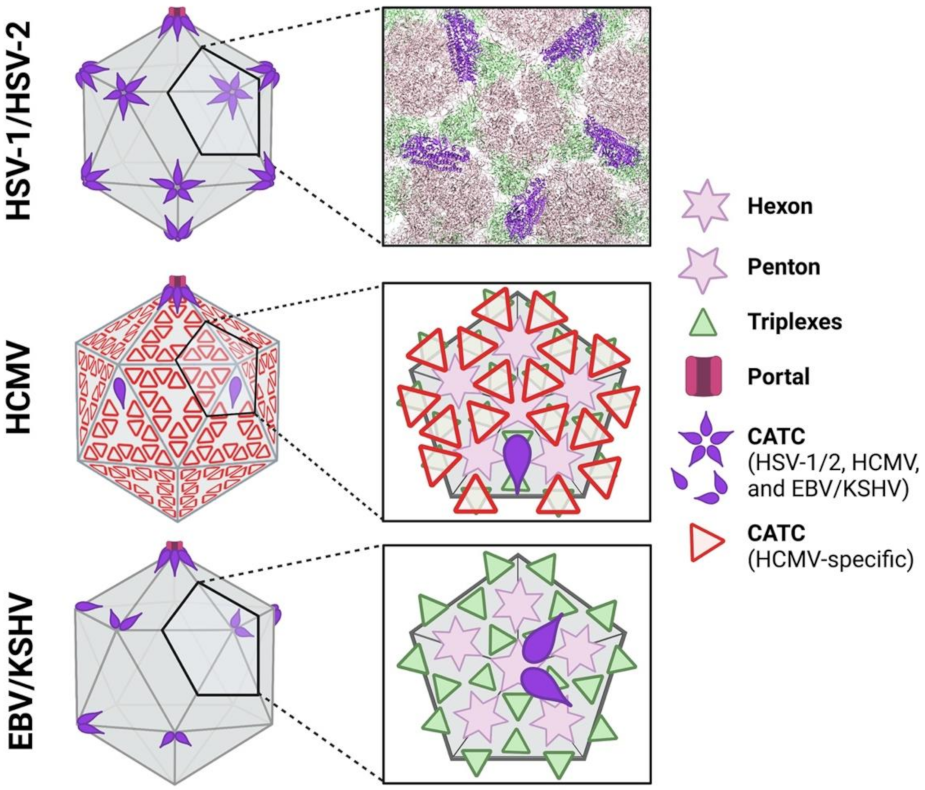
3.1.2. Functional Roles of Alphaherpesvirus CATC Proteins

3.2. Gammaherpesviruses
3.2.1. CATC Components, Capsid Location, and Occupancy
3.2.2. Functional Roles of Gammaherpesvirus CATC Proteins
3.3. Betaherpesviruses
3.3.1. CATC Components, Capsid Location, and Occupancy
3.3.2. Functional Roles of Betaherpesvirus-Specific CATC Proteins
3.3.3. Functional Roles of the Betaherpesvirus CATC Proteins Conserved in Alpha- and Gammaherpesviruses
4. Conclusions and Future Directions
- Can alternative, gentler centrifugation strategies be developed to isolate larger quantities of intact virions and capsids for in-depth studies?
- What triggers besides protease-mediated scaffold cleavage are required for DNA packaging in infected cells?
- How much does the size of the genome influence the internal capsid pressure, and does this influence CATC occupancy?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azab, W.; Osteirrieder, K. Initial Contact: The First Steps in Herpesvirus Entry. In Adv. Anat. Embryol. Cell. Biol.; Osteirrieder, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 223, pp. 1–27. [Google Scholar]
- Weidner-Glunde, M.; Kruminis-Kaszkiel, E.; Savanagouder, M. Herpesviral Latency-Common Themes. Pathogens 2020, 9, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mettenleiter, T.C. Breaching the Barrier-the Nuclear Envelope in Virus Infection. J. Mol. Biol. 2016, 428, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Roller, R.J.; Baines, J.D. Herpesvirus Nuclear Egress. In Advances in Anatomy, Embryology and Cell Biology; Osteirrieder, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 223, pp. 143–169. [Google Scholar]
- Draganova, E.B.; Thorsen, M.K.; Heldwein, E.E. Nuclear Egress. Curr. Issues Mol. Biol. 2020, 41, 125–170. [Google Scholar] [CrossRef]
- Heming, J.D.; Conway, J.F.; Homa, F.L. Herpesvirus Capsid Assembly and DNA Packaging. Adv. Anat. Embryol. Cell. Biol. 2017, 223, 119–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.C.; Newcomb, W.W. Herpesvirus Capsid Assembly: Insights from Structural Analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Zheng, Q.; Pan, D.; Yu, H.; Fu, W.; Liu, J.; He, M.; Zhu, R.; Cai, Y.; Huang, Y.; et al. Near-Atomic Cryo-Electron Microscopy Structures of Varicella-Zoster Virus Capsids. Nat. Microbiol. 2020, 5, 1542–1552. [Google Scholar] [CrossRef]
- Liu, W.; Cui, Y.; Wang, C.; Li, Z.; Gong, D.; Dai, X.; Bi, G.Q.; Sun, R.; Zhou, Z.H. Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus. Nat. Microbiol. 2020, 5, 1285–1298. [Google Scholar] [CrossRef]
- Gong, D.; Dai, X.; Jih, J.; Liu, Y.T.; Bi, G.Q.; Sun, R.; Zhou, Z.H. DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus Kshv. Cell 2019, 178, 1329–1343.e1312. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, W.; Li, Z.; Kumar, V.; Alvarez-Cabrera, A.L.; Leibovitch, E.C.; Cui, Y.; Mei, Y.; Bi, G.Q.; Jacobson, S.; et al. Atomic Structure of the Human Herpesvirus 6b Capsid and Capsid-Associated Tegument Complexes. Nat. Commun. 2019, 10, 5346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Zhou, Z.H. Structure of the Herpes Simplex Virus 1 Capsid with Associated Tegument Protein Complexes. Science 2018, 360, eaao7298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Jih, J.; Jiang, J.; Zhou, Z.H. Atomic Structure of the Human Cytomegalovirus Capsid with Its Securing Tegument Layer of Pp150. Science 2017, 356, eaam6892. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yuan, S.; Zhu, D.; Tang, H.; Wang, N.; Chen, W.; Gao, Q.; Li, Y.; Wang, J.; Liu, H.; et al. Structure of the Herpes Simplex Virus Type 2 C-Capsid with Capsid-Vertex-Specific Component. Nat. Commun. 2018, 9, 3668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhang, X.; Dong, L.; Pang, J.; Xu, M.; Zhong, Q.; Zeng, M.S.; Yu, X. Cryoem Structure of the Tegumented Capsid of Epstein-Barr Virus. Cell Res. 2020, 30, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pang, J.; Dong, L.; Yu, X. Structural Basis for Genome Packaging, Retention, and Ejection in Human Cytomegalovirus. Nat. Commun. 2021, 12, 4538. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Alain, S.; Baumert, T.F.; Ligat, G.; Hantz, S. Structures and Divergent Mechanisms in Capsid Maturation and Stabilization Following Genome Packaging of Human Cytomegalovirus and Herpesviruses. Life 2021, 11, 150. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, D.R.; Newcomb, W.W.; Brown, J.C.; Homa, F.L. Assembly of the Herpes Simplex Virus Capsid: Requirement for the Carboxyl-Terminal Twenty-Five Amino Acids of the Proteins Encoded by the Ul26 and Ul26.5 Genes. J. Virol. 1995, 69, 3690–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomsen, D.R.; Roof, L.L.; Homa, F.L. Assembly of Herpes Simplex Virus (Hsv) Intermediate Capsids in Insect Cells Infected with Recombinant Baculoviruses Expressing Hsv Capsid Proteins. J. Virol. 1994, 68, 2442–2457. [Google Scholar] [CrossRef] [Green Version]
- Tatman, J.D.; Preston, V.G.; Nicholson, P.; Elliott, R.M.; Rixon, F.J. Assembly of Herpes Simplex Virus Type 1 Capsids Using a Panel of Recombinant Baculoviruses. J. Gen. Virol. 1994, 75 Pt 5, 1101–1113. [Google Scholar] [CrossRef]
- Henson, B.W.; Perkins, E.M.; Cothran, J.E.; Desai, P. Self-Assembly of Epstein-Barr Virus Capsids. J. Virol. 2009, 83, 3877–3890. [Google Scholar] [CrossRef] [Green Version]
- Perkins, E.M.; Anacker, D.; Davis, A.; Sankar, V.; Ambinder, R.F.; Desai, P. Small Capsid Protein Porf65 Is Essential for Assembly of Kaposi’s Sarcoma-Associated Herpesvirus Capsids. J. Virol. 2008, 82, 7201–7211. [Google Scholar] [CrossRef] [Green Version]
- Newcomb, W.W.; Homa, F.L.; Thomsen, D.R.; Ye, Z.; Brown, J.C. Cell-Free Assembly of the Herpes Simplex Virus Capsid. J. Virol. 1994, 68, 6059–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newcomb, W.W.; Homa, F.L.; Thomsen, D.R.; Booy, F.P.; Trus, B.L.; Steven, A.C.; Spencer, J.V.; Brown, J.C. Assembly of the Herpes Simplex Virus Capsid: Characterization of Intermediates Observed During Cell-Free Capsid Formation. J. Mol. Biol. 1996, 263, 432–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newcomb, W.W.; Homa, F.L.; Thomsen, D.R.; Trus, B.L.; Cheng, N.; Steven, A.; Booy, F.; Brown, J.C. Assembly of the Herpes Simplex Virus Procapsid from Purified Components and Identification of Small Complexes Containing the Major Capsid and Scaffolding Proteins. J. Virol. 1999, 73, 4239–4250. [Google Scholar] [CrossRef] [Green Version]
- Newcomb, W.W.; Homa, F.L.; Brown, J.C. Involvement of the Portal at an Early Step in Herpes Simplex Virus Capsid Assembly. J. Virol. 2005, 79, 10540–10546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aksyuk, A.A.; Newcomb, W.W.; Cheng, N.; Winkler, D.C.; Fontana, J.; Heymann, J.B.; Steven, A.C. Subassemblies and Asymmetry in Assembly of Herpes Simplex Virus Procapsid. mBio 2015, 6, e01525-15. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, P.; Wang, N.; Chen, Z.; Su, D.; Zhou, Z.H.; Rao, Z.; Wang, X. Architecture of the Herpesvirus Genome-Packaging Complex and Implications for DNA Translocation. Protein Cell 2020, 11, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Homa, F.L.; Brown, J.C. Capsid Assembly and DNA Packaging in Herpes Simplex Virus. Rev. Med. Virol. 1997, 7, 107–122. [Google Scholar] [CrossRef]
- Newcomb, W.W.; Trus, B.L.; Cheng, N.; Steven, A.C.; Sheaffer, A.K.; Tenney, D.J.; Weller, S.K.; Brown, J.C. Isolation of Herpes Simplex Virus Procapsids from Cells Infected with a Protease-Deficient Mutant Virus. J. Virol. 2000, 74, 1663–1673. [Google Scholar] [CrossRef] [Green Version]
- Heymann, J.B.; Cheng, N.; Newcomb, W.W.; Trus, B.L.; Brown, J.C.; Steven, A.C. Dynamics of Herpes Simplex Virus Capsid Maturation Visualized by Time-Lapse Cryo-Electron Microscopy. Nat. Struct. Biol. 2003, 10, 334–341. [Google Scholar] [CrossRef]
- Sheaffer, A.K.; Newcomb, W.W.; Gao, M.; Yu, D.; Weller, S.K.; Brown, J.C.; Tenney, D.J. Herpes Simplex Virus DNA Cleavage and Packaging Proteins Associate with the Procapsid Prior to Its Maturation. J. Virol. 2001, 75, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Ruhge, L.L.; Huet, A.G.E.; Conway, J.F.; Smith, G.A. The Apical Region of the Herpes Simplex Virus Major Capsid Protein Promotes Capsid Maturation. J. Virol. 2018, 92, e00821-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligat, G.; Cazal, R.; Hantz, S.; Alain, S. The Human Cytomegalovirus Terminase Complex as an Antiviral Target: A Close-up View. FEMS Microbiol. Rev. 2018, 42, 137–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, D.; Weller, S.K. Herpes Simplex Virus Type 1 Cleavage and Packaging Proteins Ul15 and Ul28 Are Associated with B but Not C Capsids During Packaging. J. Virol. 1998, 72, 7428–7439. [Google Scholar] [CrossRef] [Green Version]
- Taus, N.S.; Baines, J.D. Herpes Simplex Virus 1 DNA Cleavage/Packaging: The Ul28 Gene Encodes a Minor Component of B Capsids. Virology 1998, 252, 443–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beard, P.M.; Duffy, C.; Baines, J.D. Quantification of the DNA Cleavage and Packaging Proteins U(L)15 and U(L)28 in a and B Capsids of Herpes Simplex Virus Type 1. J. Virol. 2004, 78, 1367–1374. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.C.; Baines, J.D. Herpesviruses Remodel Host Membranes for Virus Egress. Nat. Rev. Microbiol. 2011, 9, 382–394. [Google Scholar] [CrossRef]
- Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Nuclear Envelope Breakdown Can Substitute for Primary Envelopment-Mediated Nuclear Egress of Herpesviruses. J. Virol. 2011, 85, 8285–8292. [Google Scholar] [CrossRef] [Green Version]
- Desai, P.; DeLuca, N.A.; Glorioso, J.C.; Person, S. Mutations in Herpes Simplex Virus Type 1 Genes Encoding Vp5 and Vp23 Abrogate Capsid Formation and Cleavage of Replicated DNA. J. Virol. 1993, 67, 1357–1364. [Google Scholar] [CrossRef] [Green Version]
- Person, S.; Desai, P. Capsids Are Formed in a Mutant Virus Blocked at the Maturation Site of the Ul26 and Ul26.5 Open Reading Frames of Herpes Simplex Virus Type 1 but Are Not Formed in a Null Mutant of Ul38 (Vp19c). Virology 1998, 242, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional Profiling of a Human Cytomegalovirus Genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef] [Green Version]
- Desai, P.; DeLuca, N.A.; Person, S. Herpes Simplex Virus Type 1 Vp26 Is Not Essential for Replication in Cell Culture but Influences Production of Infectious Virus in the Nervous System of Infected Mice. Virology 1998, 247, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Yu, X.; Gong, H.; Jiang, X.; Abenes, G.; Liu, H.; Shivakoti, S.; Britt, W.J.; Zhu, H.; Liu, F.; et al. The Smallest Capsid Protein Mediates Binding of the Essential Tegument Protein Pp150 to Stabilize DNA-Containing Capsids in Human Cytomegalovirus. PLoS Pathog. 2013, 9, e1003525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Gong, D.; Xiao, Y.; Wu, T.T.; Sun, R.; Zhou, Z.H. Cryoem and Mutagenesis Reveal That the Smallest Capsid Protein Cements and Stabilizes Kaposi’s Sarcoma-Associated Herpesvirus Capsid. Proc. Natl. Acad. Sci. USA 2015, 112, E649–E656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathish, N.; Yuan, Y. Functional Characterization of Kaposi’s Sarcoma-Associated Herpesvirus Small Capsid Protein by Bacterial Artificial Chromosome-Based Mutagenesis. Virology 2010, 407, 306–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheaffer, A.K.; Newcomb, W.W.; Brown, J.C.; Gao, M.; Weller, S.K.; Tenney, D.J. Evidence for Controlled Incorporation of Herpes Simplex Virus Type 1 Ul26 Protease into Capsids. J. Virol. 2000, 74, 6838–6848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loveland, A.N.; Nguyen, N.L.; Brignole, E.J.; Gibson, W. The Amino-Conserved Domain of Human Cytomegalovirus Ul80a Proteins Is Required for Key Interactions During Early Stages of Capsid Formation and Virus Production. J. Virol. 2007, 81, 620–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Matusick-Kumar, L.; Hurlburt, W.; DiTusa, S.F.; Newcomb, W.W.; Brown, J.C.; McCann, P.J., 3rd; Deckman, I.; Colonno, R.J. The Protease of Herpes Simplex Virus Type 1 Is Essential for Functional Capsid Formation and Viral Growth. J. Virol. 1994, 68, 3702–3712. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Trang, P.; Shah, S.; Atanasov, I.; Kim, Y.H.; Bai, Y.; Zhou, Z.H.; Liu, F. Dissecting Human Cytomegalovirus Gene Function and Capsid Maturation by Ribozyme Targeting and Electron Cryomicroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 7103–7108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsurumi, S.; Watanabe, T.; Iwaisako, Y.; Suzuki, Y.; Nakano, T.; Fujimuro, M. Kaposi’s Sarcoma-Associated Herpesvirus Orf17 Plays a Key Role in Capsid Maturation. Virology 2021, 558, 76–85. [Google Scholar] [CrossRef]
- Desai, P.; Watkins, S.C.; Person, S. The Size and Symmetry of B Capsids of Herpes Simplex Virus Type 1 Are Determined by the Gene Products of the Ul26 Open Reading Frame. J. Virol. 1994, 68, 5365–5374. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.H.; Rixon, F.J.; Cunningham, C.; Davison, A.J. Isolation and Characterization of Herpes Simplex Virus Type 1 Mutants Defective in the Ul6 Gene. Virology 1996, 217, 111–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huffman, J.B.; Newcomb, W.W.; Brown, J.C.; Homa, F.L. Amino Acids 143 to 150 of the Herpes Simplex Virus Type 1 Scaffold Protein Are Required for the Formation of Portal-Containing Capsids. J. Virol. 2008, 82, 6778–6781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Baines, J.D. Domain within Herpes Simplex Virus 1 Scaffold Proteins Required for Interaction with Portal Protein in Infected Cells and Incorporation of the Portal Vertex into Capsids. J. Virol. 2008, 82, 5021–5030. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, V.; Sommer, M.; Rajamani, J.; Zerboni, L.; Arvin, A.M. Functions of Varicella-Zoster Virus Orf23 Capsid Protein in Viral Replication and the Pathogenesis of Skin Infection. J. Virol. 2008, 82, 10231–10246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trus, B.L.; Booy, F.P.; Newcomb, W.W.; Brown, J.C.; Homa, F.L.; Thomsen, D.R.; Steven, A.C. The Herpes Simplex Virus Procapsid: Structure, Conformational Changes Upon Maturation, and Roles of the Triplex Proteins Vp19c and Vp23 in Assembly. J. Mol. Biol. 1996, 263, 447–462. [Google Scholar] [CrossRef]
- Newcomb, W.W.; Thomsen, D.R.; Homa, F.L.; Brown, J.C. Assembly of the Herpes Simplex Virus Capsid: Identification of Soluble Scaffold-Portal Complexes and Their Role in Formation of Portal-Containing Capsids. J. Virol. 2003, 77, 9862–9871. [Google Scholar] [CrossRef] [Green Version]
- Singer, G.P.; Newcomb, W.W.; Thomsen, D.R.; Homa, F.L.; Brown, J.C. Identification of a Region in the Herpes Simplex Virus Scaffolding Protein Required for Interaction with the Portal. J. Virol. 2005, 79, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Huang, E.; Perkins, E.M.; Desai, P. Structural Features of the Scaffold Interaction Domain at the N Terminus of the Major Capsid Protein (Vp5) of Herpes Simplex Virus Type 1. J. Virol. 2007, 81, 9396–9407. [Google Scholar] [CrossRef] [Green Version]
- Preston, V.G.; Coates, J.A.; Rixon, F.J. Identification and Characterization of a Herpes Simplex Virus Gene Product Required for Encapsidation of Virus DNA. J. Virol. 1983, 45, 1056–1064. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, R.; Kato, A.; Sagara, H.; Watanabe, M.; Maruzuru, Y.; Koyanagi, N.; Arii, J.; Kawaguchi, Y. Herpes Simplex Virus 1 Small Capsomere-Interacting Protein Vp26 Regulates Nucleocapsid Maturation. J. Virol. 2017, 91, e01068-17. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.H.; Jakana, J.; McNab, D.; Mitchell, J.; Zhou, Z.H.; Dougherty, M.; Chiu, W.; Rixon, F.J. The Pattern of Tegument-Capsid Interaction in the Herpes Simplex Virus Type 1 Virion Is Not Influenced by the Small Hexon-Associated Protein Vp26. J. Virol. 2001, 75, 11863–11867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luxton, G.W.; Haverlock, S.; Coller, K.E.; Antinone, S.E.; Pincetic, A.; Smith, G.A. Targeting of Herpesvirus Capsid Transport in Axons Is Coupled to Association with Specific Sets of Tegument Proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 5832–5837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzesik, P.; MacMath, D.; Henson, B.; Prasad, S.; Joshi, P.; Desai, P.J. Incorporation of the Kaposi’s Sarcoma-Associated Herpesvirus Capsid Vertex-Specific Component (Cvsc) into Self-Assembled Capsids. Virus Res. 2017, 236, 9–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borst, E.M.; Mathys, S.; Wagner, M.; Muranyi, W.; Messerle, M. Genetic Evidence of an Essential Role for Cytomegalovirus Small Capsid Protein in Viral Growth. J. Virol. 2001, 75, 1450–1458. [Google Scholar] [CrossRef] [Green Version]
- Honess, R.W. Herpes Simplex and ‘the Herpes Complex’: Diverse Observations and a Unifying Hypothesis. The Eighth Fleming Lecture. J. Gen. Virol. 1984, 65 Pt 12, 2077–2107. [Google Scholar] [CrossRef]
- Sae-Ueng, U.; Liu, T.; Catalano, C.E.; Huffman, J.B.; Homa, F.L.; Evilevitch, A. Major Capsid Reinforcement by a Minor Protein in Herpesviruses and Phage. Nucleic Acids Res. 2014, 42, 9096–9107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trus, B.L.; Newcomb, W.W.; Cheng, N.; Cardone, G.; Marekov, L.; Homa, F.L.; Brown, J.C.; Steven, A.C. Allosteric Signaling and a Nuclear Exit Strategy: Binding of Ul25/Ul17 Heterodimers to DNA-Filled Hsv-1 Capsids. Mol. Cell 2007, 26, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Toropova, K.; Huffman, J.B.; Homa, F.L.; Conway, J.F. The Herpes Simplex Virus 1 Ul17 Protein Is the Second Constituent of the Capsid Vertex-Specific Component Required for DNA Packaging and Retention. J. Virol. 2011, 85, 7513–7522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Gong, D.; Wu, T.T.; Sun, R.; Zhou, Z.H. Organization of Capsid-Associated Tegument Components in Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2014, 88, 12694–12702. [Google Scholar] [CrossRef] [Green Version]
- Homa, F.L.; Huffman, J.B.; Toropova, K.; Lopez, H.R.; Makhov, A.M.; Conway, J.F. Structure of the Pseudorabies Virus Capsid: Comparison with Herpes Simplex Virus Type 1 and Differential Binding of Essential Minor Proteins. J. Mol. Biol. 2013, 425, 3415–3428. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Dai, X.; Jih, J.; Chan, K.; Trang, P.; Yu, X.; Balogun, R.; Mei, Y.; Liu, F.; Zhou, Z.H. Atomic Structures and Deletion Mutant Reveal Different Capsid-Binding Patterns and Functional Significance of Tegument Protein Pp150 in Murine and Human Cytomegaloviruses with Implications for Therapeutic Development. PLoS Pathog. 2019, 15, e1007615. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.T.; Jih, J.; Dai, X.; Bi, G.Q.; Zhou, Z.H. Cryo-Em Structures of Herpes Simplex Virus Type 1 Portal Vertex and Packaged Genome. Nature 2019, 570, 257–261. [Google Scholar] [CrossRef]
- McElwee, M.; Vijayakrishnan, S.; Rixon, F.; Bhella, D. Structure of the Herpes Simplex Virus Portal-Vertex. PLoS Biol. 2018, 16, e2006191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Chen, W.; Zhu, L.; Zhu, D.; Feng, R.; Wang, J.; Zhu, B.; Zhang, X.; Chen, X.; Liu, X.; et al. Structures of the Portal Vertex Reveal Essential Protein-Protein Interactions for Herpesvirus Assembly and Maturation. Protein Cell 2020, 11, 366–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebrun, M.; Thelen, N.; Thiry, M.; Riva, L.; Ote, I.; Condé, C.; Vandevenne, P.; Di Valentin, E.; Bontems, S.; Sadzot-Delvaux, C. Varicella-Zoster Virus Induces the Formation of Dynamic Nuclear Capsid Aggregates. Virology 2014, 454–455, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Parvate, A.D.; Vago, F.; Hasan, S.S.; Lee, J.; Williams, E.P.; Lanman, J.; Jonsson, C.B. A New Inactivation Method to Facilitate Cryo-Em of Enveloped, Rna Viruses Requiring High Containment: A Case Study Using Venezuelan Equine Encephalitis Virus (Veev). J. Virol. Methods 2020, 277, 113792. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Salmon, B.; Cunningham, C.; Davison, A.J.; Harris, W.J.; Baines, J.D. The Herpes Simplex Virus Type 1 U(L)17 Gene Encodes Virion Tegument Proteins That Are Required for Cleavage and Packaging of Viral DNA. J. Virol. 1998, 72, 3779–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockrell, S.K.; Sanchez, M.E.; Erazo, A.; Homa, F.L. Role of the Ul25 Protein in Herpes Simplex Virus DNA Encapsidation. J. Virol. 2009, 83, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Stow, N.D. Packaging of Genomic and Amplicon DNA by the Herpes Simplex Virus Type 1 Ul25-Null Mutant Kul25ns. J. Virol. 2001, 75, 10755–10765. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, J.; Leege, T.; Klupp, B.G.; Granzow, H.; Fuchs, W.; Mettenleiter, T.C. Partial Functional Complementation of a Pseudorabies Virus Ul25 Deletion Mutant by Herpes Simplex Virus Type 1 Pul25 Indicates Overlapping Functions of Alphaherpesvirus Pul25 Proteins. J. Virol. 2008, 82, 5725–5734. [Google Scholar] [CrossRef] [Green Version]
- Klupp, B.G.; Granzow, H.; Keil, G.M.; Mettenleiter, T.C. The Capsid-Associated Ul25 Protein of the Alphaherpesvirus Pseudorabies Virus Is Nonessential for Cleavage and Encapsidation of Genomic DNA but Is Required for Nuclear Egress of Capsids. J. Virol. 2006, 80, 6235–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNab, A.R.; Desai, P.; Person, S.; Roof, L.L.; Thomsen, D.R.; Newcomb, W.W.; Brown, J.C.; Homa, F.L. The Product of the Herpes Simplex Virus Type 1 Ul25 Gene Is Required for Encapsidation but Not for Cleavage of Replicated Viral DNA. J. Virol. 1998, 72, 1060–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandariz-Nuñez, A.; Liu, T.; Du, T.; Evilevitch, A. Pressure-Driven Release of Viral Genome into a Host Nucleus Is a Mechanism Leading to Herpes Infection. Elife 2019, 8, e47212. [Google Scholar] [CrossRef] [PubMed]
- Snijder, J.; Radtke, K.; Anderson, F.; Scholtes, L.; Corradini, E.; Baines, J.; Heck, A.J.R.; Wuite, G.J.L.; Sodeik, B.; Roos, W.H. Vertex-Specific Proteins Pul17 and Pul25 Mechanically Reinforce Herpes Simplex Virus Capsids. J. Virol. 2017, 91, e00123-17. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. The Pymol Molecular Graphics System, Version 2.5.1; Schrödinger Inc.: New York, NY, USA, 2015; Available online: https://pymol.org/2/support.html (accessed on 19 September 2021).
- Cardone, G.; Newcomb, W.W.; Cheng, N.; Wingfield, P.T.; Trus, B.L.; Brown, J.C.; Steven, A.C. The Ul36 Tegument Protein of Herpes Simplex Virus 1 Has a Composite Binding Site at the Capsid Vertices. J. Virol. 2012, 86, 4058–4064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, D.J.; Crump, C.M.; Graham, S.C. Tegument Assembly and Secondary Envelopment of Alphaherpesviruses. Viruses 2015, 7, 5084–5114. [Google Scholar] [CrossRef] [Green Version]
- Luxton, G.W.; Lee, J.I.; Haverlock-Moyns, S.; Schober, J.M.; Smith, G.A. The Pseudorabies Virus Vp1/2 Tegument Protein Is Required for Intracellular Capsid Transport. J. Virol. 2006, 80, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Sandbaumhüter, M.; Döhner, K.; Schipke, J.; Binz, A.; Pohlmann, A.; Sodeik, B.; Bauerfeind, R. Cytosolic Herpes Simplex Virus Capsids Not Only Require Binding Inner Tegument Protein Pul36 but Also Pul37 for Active Transport Prior to Secondary Envelopment. Cell Microbiol. 2013, 15, 248–269. [Google Scholar] [CrossRef]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes Simplex Virus Replication: Roles of Viral Proteins and Nucleoporins in Capsid-Nucleus Attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef] [Green Version]
- Abaitua, F.; Daikoku, T.; Crump, C.M.; Bolstad, M.; O’Hare, P. A Single Mutation Responsible for Temperature-Sensitive Entry and Assembly Defects in the Vp1-2 Protein of Herpes Simplex Virus. J. Virol. 2011, 85, 2024–2036. [Google Scholar] [CrossRef] [Green Version]
- Pasdeloup, D.; Blondel, D.; Isidro, A.L.; Rixon, F.J. Herpesvirus Capsid Association with the Nuclear Pore Complex and Viral DNA Release Involve the Nucleoporin Can/Nup214 and the Capsid Protein Pul25. J. Virol. 2009, 83, 6610–6623. [Google Scholar] [CrossRef] [Green Version]
- Huffman, J.B.; Daniel, G.R.; Falck-Pedersen, E.; Huet, A.; Smith, G.A.; Conway, J.F.; Homa, F.L. The C Terminus of the Herpes Simplex Virus Ul25 Protein Is Required for Release of Viral Genomes from Capsids Bound to Nuclear Pores. J. Virol. 2017, 91, e00641-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharkwal, H.; Smith, C.G.; Wilson, D.W. Herpes Simplex Virus Capsid Localization to Escrt-Vps4 Complexes in the Presence and Absence of the Large Tegument Protein Ul36p. J. Virol. 2016, 90, 7257–7267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeshima, K.; Arii, J.; Maruzuru, Y.; Koyanagi, N.; Kato, A.; Kawaguchi, Y. Identification of the Capsid Binding Site in the Herpes Simplex Virus 1 Nuclear Egress Complex and Its Role in Viral Primary Envelopment and Replication. J. Virol. 2019, 93, e01290-19. [Google Scholar] [CrossRef] [PubMed]
- Draganova, E.B.; Zhang, J.; Zhou, Z.H.; Heldwein, E.E. Structural Basis for Capsid Recruitment and Coat Formation During Hsv-1 Nuclear Egress. Elife 2020, 9, e56627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Selariu, A.; Warden, C.; Huang, G.; Huang, Y.; Zaccheus, O.; Cheng, T.; Xia, N.; Zhu, H. Genome-Wide Mutagenesis Reveals That Orf7 Is a Novel Vzv Skin-Tropic Factor. PLoS Pathog. 2010, 6, e1000971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Dünn-Kittenplon, D.; Ashkenazy-Titelman, A.; Kalt, I.; Lellouche, J.P.; Shav-Tal, Y.; Sarid, R. The Portal Vertex of Kshv Promotes Docking of Capsids at the Nuclear Pores. Viruses 2021, 13, 597. [Google Scholar] [CrossRef]
- Dolan, A.; Cunningham, C.; Hector, R.D.; Hassan-Walker, A.F.; Lee, L.; Addison, C.; Dargan, D.J.; McGeoch, D.J.; Gatherer, D.; Emery, V.C.; et al. Genetic Content of Wild-Type Human Cytomegalovirus. J. Gen. Virol. 2004, 85, 1301–1312. [Google Scholar] [CrossRef]
- Dominguez, G.; Dambaugh, T.R.; Stamey, F.R.; Dewhurst, S.; Inoue, N.; Pellett, P.E. Human Herpesvirus 6b Genome Sequence: Coding Content and Comparison with Human Herpesvirus 6a. J. Virol. 1999, 73, 8040–8052. [Google Scholar] [CrossRef] [Green Version]
- McGeoch, D.J.; Dalrymple, M.A.; Davison, A.J.; Dolan, A.; Frame, M.C.; McNab, D.; Perry, L.J.; Scott, J.E.; Taylor, P. The Complete DNA Sequence of the Long Unique Region in the Genome of Herpes Simplex Virus Type 1. J. Gen. Virol. 1988, 69 Pt 7, 1531–1574. [Google Scholar] [CrossRef]
- Nicholas, J. Determination and Analysis of the Complete Nucleotide Sequence of Human Herpesvirus. J. Virol. 1996, 70, 5975–5989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhella, D.; Rixon, F.J.; Dargan, D.J. Cryomicroscopy of Human Cytomegalovirus Virions Reveals More Densely Packed Genomic DNA Than in Herpes Simplex Virus Type 1. J. Mol. Biol. 2000, 295, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Mocarski, E.S. Cytomegalovirus Pul96 Is Critical for the Stability of Pp150-Associated Nucleocapsids. J. Virol. 2011, 85, 7129–7141. [Google Scholar] [CrossRef] [Green Version]
- Indran, S.V.; Ballestas, M.E.; Britt, W.J. Bicaudal D1-Dependent Trafficking of Human Cytomegalovirus Tegument Protein Pp150 in Virus-Infected Cells. J. Virol. 2010, 84, 3162–3177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AuCoin, D.P.; Smith, G.B.; Meiering, C.D.; Mocarski, E.S. Betaherpesvirus-Conserved Cytomegalovirus Tegument Protein Ppul32 (Pp150) Controls Cytoplasmic Events During Virion Maturation. J. Virol. 2006, 80, 8199–8210. [Google Scholar] [CrossRef] [Green Version]
- Bogdanow, B.; Weisbach, H.; von Einem, J.; Straschewski, S.; Voigt, S.; Winkler, M.; Hagemeier, C.; Wiebusch, L. Human Cytomegalovirus Tegument Protein Pp150 Acts as a Cyclin A2-Cdk-Dependent Sensor of the Host Cell Cycle and Differentiation State. Proc. Natl. Acad. Sci. USA 2013, 110, 17510–17515. [Google Scholar] [CrossRef] [Green Version]
- Borst, E.M.; Bauerfeind, R.; Binz, A.; Stephan, T.M.; Neuber, S.; Wagner, K.; Steinbrück, L.; Sodeik, B.; Roviš, T.L.; Jonjić, S.; et al. The Essential Human Cytomegalovirus Proteins Pul77 and Pul93 Are Structural Components Necessary for Viral Genome Encapsidation. J. Virol. 2016, 90, 5860–5875. [Google Scholar] [CrossRef] [Green Version]
- DeRussy, B.M.; Tandon, R. Human Cytomegalovirus Pul93 Is Required for Viral Genome Cleavage and Packaging. J. Virol. 2015, 89, 12221–12225. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.E.; Oh, S.E.; Kwon, K.M.; Lee, C.H.; Ahn, J.H. Involvement of the N-Terminal Deubiquitinating Protease Domain of Human Cytomegalovirus Ul48 Tegument Protein in Autoubiquitination, Virion Stability, and Virus Entry. J. Virol. 2016, 90, 3229–3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Ortiz, D.A.; Gurczynski, S.J.; Khan, F.; Pellett, P.E. Identification of Human Cytomegalovirus Genes Important for Biogenesis of the Cytoplasmic Virion Assembly Complex. J. Virol. 2014, 88, 9086–9099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brock, I.; Krüger, M.; Mertens, T.; von Einem, J. Nuclear Targeting of Human Cytomegalovirus Large Tegument Protein Pul48 Is Essential for Viral Growth. J. Virol. 2013, 87, 6005–6019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Shah, S.; Lee, M.; Dai, W.; Lo, P.; Britt, W.; Zhu, H.; Liu, F.; Zhou, Z.H. Biochemical and Structural Characterization of the Capsid-Bound Tegument Proteins of Human Cytomegalovirus. J. Struct. Biol. 2011, 174, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Inoue, N.; Matsushita, M.; Fukui, Y.; Yamada, S.; Tsuda, M.; Higashi, C.; Kaneko, K.; Hasegawa, H.; Yamaguchi, T. Identification of a Varicella-Zoster Virus Replication Inhibitor That Blocks Capsid Assembly by Interacting with the Floor Domain of the Major Capsid Protein. J. Virol. 2012, 86, 12198–12207. [Google Scholar] [CrossRef] [Green Version]
- Keil, T.; Liu, D.; Lloyd, M.; Coombs, W.; Moffat, J.; Visalli, R. DNA Encapsidation and Capsid Assembly Are Underexploited Antiviral Targets for the Treatment of Herpesviruses. Front. Microbiol. 2020, 11, 1862. [Google Scholar] [CrossRef]

| Alpha | Beta | Gamma | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSV-1 | HSV-2 | VZV | HCMV | HHV-6 | EBV | KSHV | ||||||||
| Gene | Protein | Gene | Protein | Gene | Protein | Gene | Protein | Gene | Protein | Gene | Protein | Gene | Protein | |
| Portal | UL6 | UL6 | UL6 | UL6 | ORF54 | ORF54 | UL104 | Portal | U76 | Portal | BBRF1 | BBRF1 | ORF43 | ORF43 |
| MCP | UL19 | VP5 | UL19 | VP5 | ORF40 | ORF40 | UL86 | MCP | U57 | MCP | BcLF1 | BcLF1 | ORF25 | ORF25 |
| SCP | UL35 | VP26 | UL35 | VP26 | ORF23 | ORF23 | UL48a (UL48.5) | SCP | U53 | SCP | BVRF3 | BFRF3 | ORF65 | ORF65 |
| Triplex 1 | UL38 | VP19c | UL38 | VP19c | ORF20 | ORF20 | UL46 | mCP-BP | U29 | Triplex 1 | BORF1 | BORF1 | ORF62 | ORF62 |
| Triplex 2 | UL18 | VP23 | UL18 | VP23 | ORF41 | ORF41 | UL85 | mCP | U56 | Triplex 2 | BDLF1 | BDLF1 | ORF26 | ORF26 |
| CATC | UL25 | UL25 | UL25 | UL25 | ORF34 | ORF34 | UL77 | UL77 | U50 | U50 * | BVRF1 | CVC2 | ORF19 | ORF19 |
| UL17 | UL17 | UL17 | UL17 | ORF43 | ORF43 | UL93 | UL93 | U64 | U64 * | BGLF1 | CVC1 | ORF32 | ORF32 | |
| UL36 | UL36 | UL36 | UL36 | ORF22 | ORF22 | UL48 | UL48 | U31 | U31 * | BPLF1 | LTP | ORF64 | ORF64 | |
| CATC (beta only) | -- | -- | -- | -- | -- | -- | UL32 | pp150 | U11 | U11 | -- | -- | -- | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Draganova, E.B.; Valentin, J.; Heldwein, E.E. The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies. Viruses 2021, 13, 1913. https://doi.org/10.3390/v13101913
Draganova EB, Valentin J, Heldwein EE. The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies. Viruses. 2021; 13(10):1913. https://doi.org/10.3390/v13101913
Chicago/Turabian StyleDraganova, Elizabeth B., Jonathan Valentin, and Ekaterina E. Heldwein. 2021. "The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies" Viruses 13, no. 10: 1913. https://doi.org/10.3390/v13101913
APA StyleDraganova, E. B., Valentin, J., & Heldwein, E. E. (2021). The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies. Viruses, 13(10), 1913. https://doi.org/10.3390/v13101913






