BAF45b Is Required for Efficient Zika Virus Infection of HAP1 Cells
Abstract
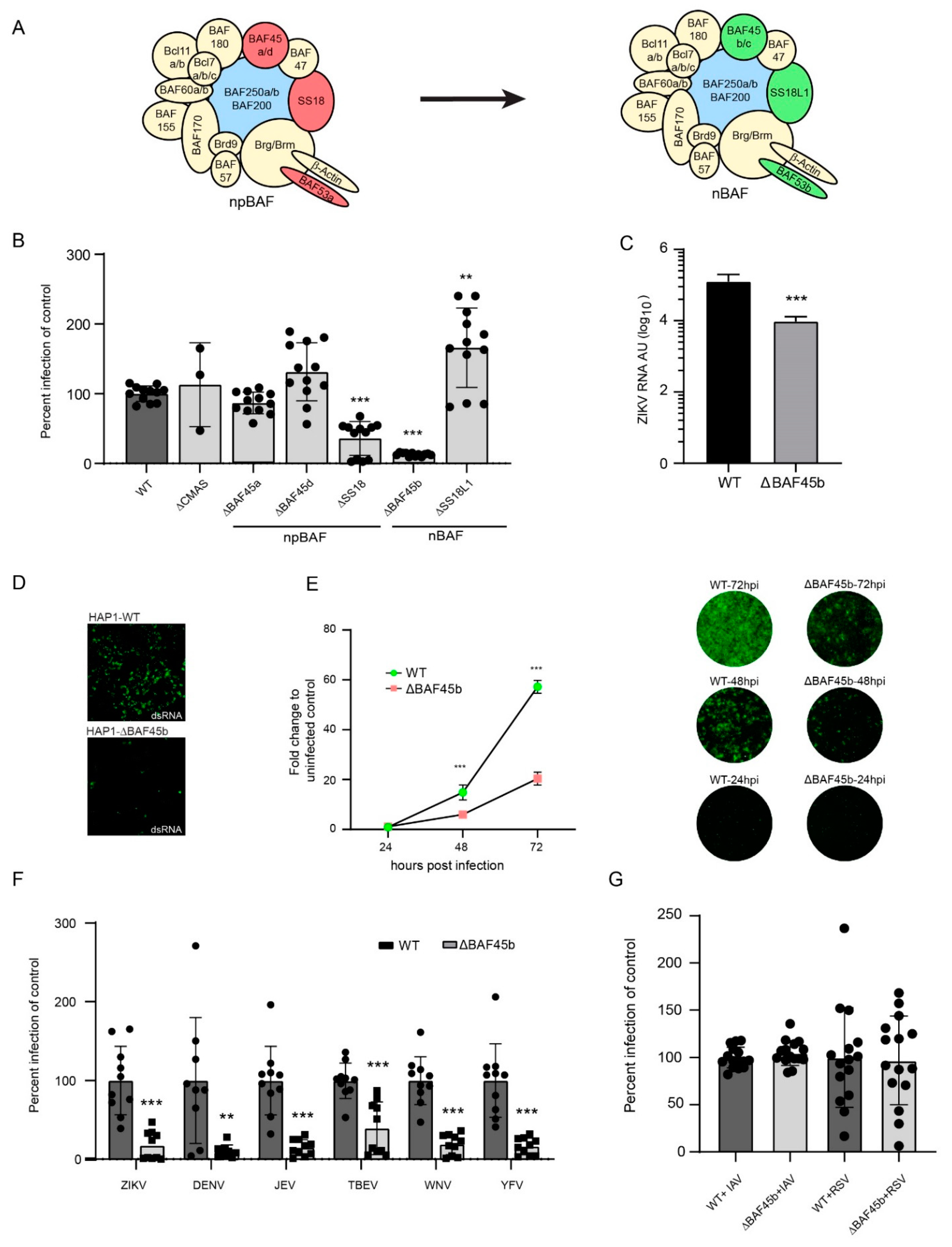
:1. Introduction
2. Materials and Methods
2.1. Viruses, Antibodies, Reagents and Cells
2.2. NCI-60 Screen for ZIKV Infection
2.3. Combined DAVID Functional Annotation Clustering & STRING Interactions
2.4. Quantification of ZIKV Infection in HAP1-WT/HAP1-KO Cells by Immunofluorescence
2.5. Quantification of ZIKV Infection by qRT-PCR
2.6. Infection of HAP1 Cells by Flavivirus, RSV, and IAV
2.7. Statistical Analysis
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Heymann, D.L.; Hodgson, A.; Sall, A.A.; Freedman, D.O.; Staples, J.E.; Althabe, F.; Baruah, K.; Mahmud, G.; Kandun, N.; Vasconcelos, P.F.C.; et al. Zika virus and microcephaly: Why is this situation a PHEIC? Lancet 2016, 387, 719–721. [Google Scholar] [CrossRef] [Green Version]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.; Zogbi, H.E.; Calvet, G.; de Souza, R.V.; Siqueira, A.; de Mendonca, M.C.L.; Nogueira, R.M.R.; de Filippis, A.M.B.; et al. Guillain-Barré syndrome associated with Zika virus infection. Lancet 2016, 387, 1482. [Google Scholar] [CrossRef] [Green Version]
- Samarasekera, U.; Triunfol, M. Concern over Zika virus grips the world. Lancet 2016, 387, 521–524. [Google Scholar] [CrossRef] [Green Version]
- PAHO/WHO. Zika—Epidemiological Report Brazil. Available online: https://www.paho.org/hq/dmdocuments/2017/2017-phe-zika-situation-report-bra.pdf (accessed on 25 September 2017).
- Boorman, J.P.T.; Porterfield, J.S. A simple technique for infection of mosquitoes with viruses transmission of Zika virus. Trans. R. Soc. Trop. Med. Hyg. 1956, 50, 238–242. [Google Scholar] [CrossRef]
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef] [Green Version]
- Chimelli, L.; Melo, A.S.O.; Avvad-Portari, E.; Wiley, C.A.; Camacho, A.H.S.; Lopes, V.S.; Machado, H.N.; Andrade, C.V.; Dock, D.C.A.; Moreira, M.E.; et al. The spectrum of neuropathological changes associated with congenital Zika virus infection. Acta Neuropathol. 2017, 133, 983–999. [Google Scholar] [CrossRef]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, F.B.R.G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.-F.; Xu, Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 2016, 19, 120–126. [Google Scholar] [CrossRef] [Green Version]
- Pierson, T.C.; Diamond, M.S. Flaviviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Roizman, B., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 747–794. [Google Scholar]
- Zhang, H.; Chang, Y.; Zhang, L.; Kim, S.-N.; Otaegi, G.; Zhang, Z.; Nie, Y.; Mubarak, T.; Li, C.; Qin, C.-F.; et al. Upregulation of MicroRNA miR-9 Is Associated with Microcephaly and Zika Virus Infection in Mice. Mol. Neurobiol. 2019, 56, 4072–4085. [Google Scholar] [CrossRef]
- Rashid, M.-U.; Zahedi-Amiri, A.; Glover, K.K.M.; Gao, A.; Nickol, M.E.; Kindrachuk, J.; Wilkins, J.A.; Coombs, K.M. Zika virus dysregulates human Sertoli cell proteins involved in spermatogenesis with little effect on tight junctions. PLoS Negl. Trop. Dis. 2020, 14, e0008335. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Acklin, J.A.; Liu, G.; Kenney, H.; Teterina, N.L.; Pletnev, A.G.; Lim, J.K. Zika virus tropism during early infection of the testicular interstitium and its role in viral pathogenesis in the testes. PLoS Pathog. 2020, 16, e1008601. [Google Scholar] [CrossRef]
- Azouz, F.; Arora, K.; Krause, K.; Nerurkar, V.R.; Kumar, M. Integrated MicroRNA and mRNA Profiling in Zika Virus-Infected Neurons. Viruses 2019, 11, 162. [Google Scholar] [CrossRef] [Green Version]
- Dang, J.W.; Tiwari, S.K.; Qin, Y.; Rana, T.M. Genome-wide Integrative Analysis of Zika-Virus-Infected Neuronal Stem Cells Reveals Roles for MicroRNAs in Cell Cycle and Stemness. Cell Rep. 2019, 27, 3618–3628. [Google Scholar] [CrossRef] [Green Version]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [Green Version]
- Sousa, S.B.; Hennekam, R.C.; The Nicolaides–Baraitser Syndrome International Consortium. Phenotype and genotype in Nicolaides-Baraitser syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166C, 302–314. [Google Scholar] [CrossRef]
- Tsurusaki, Y.; Koshimizu, E.; Ohashi, H.; Phadke, S.; Kou, I.; Shiina, M.; Suzuki, T.; Okamoto, N.; Imamura, S.; Yamashita, M.; et al. De novo SOX11 mutations cause Coffin-Siris syndrome. Nat. Commun. 2014, 5, 4011. [Google Scholar] [CrossRef] [Green Version]
- Dias, C.; Estruch, S.B.; Graham, S.; McRae, J.; Sawiak, S.; Hurst, J.A.; Joss, S.K.; Holder, S.E.; Morton, J.E.; Turner, C.; et al. BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. Am. J. Hum. Genet. 2016, 99, 253–274. [Google Scholar] [CrossRef] [Green Version]
- Son, E.Y.; Crabtree, G.R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 333–349. [Google Scholar] [CrossRef] [Green Version]
- Sokpor, G.; Xie, Y.; Rosenbusch, J.; Tuoc, T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci. 2017, 10, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenderski, W.; Wang, L.; Krokhotin, A.; Walsh, J.J.; Li, H.; Shoji, H.; Ghosh, S.; George, R.D.; Miller, E.L.; Elias, L.; et al. Loss of the neural-specific BAF subunit ACTL6B relieves repression of early response genes and causes recessive autism. Proc. Natl. Acad. Sci. USA 2020, 117, 10055–10066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saladi, S.; Ross, K.; Karaayvaz, M.; Tata, P.R.; Mou, H.; Rajagopal, J.; Ramaswamy, S.; Ellisen, L.W. ACTL6A Is Co-Amplified with p63 in Squamous Cell Carcinoma to Drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell 2017, 31, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, G.; Paul, S.; Beshara, S.; Ramanujan, V.K.; Ramaiah, A.; Nielsen-Saines, K.; Li, M.M.; French, S.W.; Morizono, K.; Kumar, A.; et al. Hippo Signaling Pathway Has a Critical Role in Zika Virus Replication and in the Pathogenesis of Neuroinflammation. Am. J. Pathol. 2020, 190, 844–861. [Google Scholar] [CrossRef] [PubMed]
- Kandilya, D.; Maskomani, S.; Shyamasundar, S.; Tambyah, P.A.; Yng, C.S.; Lee, R.C.H.; Hande, M.P.; Mallilankaraman, K.; Chu, J.J.H.; Dheen, S.T. Zika virus alters DNA methylation status of genes involved in Hippo signaling pathway in human neural progenitor cells. Epigenomics 2019, 11, 1143–1161. [Google Scholar] [CrossRef]
- Savidis, G.; McDougall, W.M.; Meraner, P.; Perreira, J.M.; Portmann, J.M.; Trincucci, G.; John, S.P.; Aker, A.M.; Renzette, N.; Robbins, D.R.; et al. Identification of Zika Virus and Dengue Virus Dependency Factors Using Functional Genomics. Cell Rep. 2016, 16, 232–246. [Google Scholar] [CrossRef] [Green Version]
- Scaturro, P.; Stukalov, A.; Haas, D.; Cortese, M.; Draganova, K.; Płaszczyca, A.; Bartenschlager, R.; Götz, M.; Pichlmair, A. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 2018, 561, 253–257. [Google Scholar] [CrossRef]
- Donald, C.L.; Brennan, B.; Cumberworth, S.L.; Rezelj, V.V.; Clark, J.J.; Cordeiro, M.T.; Freitas de Oliveira França, R.; Pena, L.J.; Wilkie, G.S.; Da Silva Filipe, A.; et al. Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0005048. [Google Scholar] [CrossRef]
- Asghar, N.; Lee, Y.-P.; Nilsson, E.; Lindqvist, R.; Melik, W.; Kröger, A.; Överby, A.K.; Johansson, M. The role of the poly(A) tract in the replication and virulence of tick-borne encephalitis virus. Sci. Rep. 2016, 6, 39265. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, R.; Mundt, F.; Gilthorpe, J.D.; Wölfel, S.; Gekara, N.O.; Kröger, A.; Överby, A.K. Fast type I interferon response protects astrocytes from flavivirus infection and virus-induced cytopathic effects. J. Neuroinflamm. 2016, 13, 277. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, J.; Snijder, B.; Sacher, R.; Burkard, C.; Bleck, C.; Stahlberg, H.; Pelkmans, L.; Helenius, A. RNAi Screening Reveals Proteasome- and Cullin3-Dependent Stages in Vaccinia Virus Infection. Cell Rep. 2012, 2, 1036–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NIH-NCI. US National Cancer Institute: The NCI-60 Human Tumor Cell Lines Screen. Available online: https://dtp.cancer.gov/discovery_development/nci-60/default.htm (accessed on 11 August 2020).
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Di Pasquale, G.; Davidson, B.L.; Stein, C.S.; Martins, I.; Scudiero, D.; Monks, A.; Chiorini, J.A. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 2003, 9, 1306–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chestkov, A.V.; Baka, I.D.; Kost, M.V.; Georgiev, G.P.; Buchman, V.L. The d4 Gene Family in the Human Genome. Genomics 1996, 36, 174–177. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- The Human Protein Atlas. DPF1. Available online: https://www.proteinatlas.org/ENSG00000011332-DPF1 (accessed on 24 February 2021).
- Tsetsarkin, K.A.; Maximova, O.A.; Liu, G.; Kenney, H.; Teterina, N.; Bloom, M.E.; Grabowski, J.M.; Mlera, L.; Nagata, B.M.; Moore, I.; et al. Routes of Zika virus dissemination in the testis and epididymis of immunodeficient mice. Nat. Commun. 2018, 9, 5350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carette, J.; Raaben, M.; Wong, A.C.; Herbert, A.S.; Obernosterer, G.; Mulherkar, N.; Kuehne, A.I.; Kranzusch, P.J.; Griffin, A.M.; Ruthel, G.; et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011, 477, 340–343. [Google Scholar] [CrossRef] [Green Version]
- Schick, S.; Rendeiro, A.F.; Runggatscher, K.; Ringler, A.; Boidol, B.; Hinkel, M.; Májek, P.; Vulliard, L.; Penz, T.; Parapatics, K.; et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat. Genet. 2019, 51, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Akbar, I.; Bhagat, R.; Hazra, B.; Bhattacharyya, A.; Seth, P.; Roy, D.; Basu, A. Identification and Classification of Hubs in microRNA Target Gene Networks in Human Neural Stem/Progenitor Cells following Japanese Encephalitis Virus Infection. mSphere 2019, 4, e00588-19. [Google Scholar] [CrossRef] [Green Version]
- Marceau, C.D.; Puschnik, A.S.; Majzoub, K.; Ooi, Y.S.; Brewer, S.M.; Fuchs, G.; Swaminathan, K.; Mata, M.A.; Elias, J.E.; Sarnow, P.; et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535, 159–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Dong, X.; Li, S.-H.; Zhou, Y.-P.; Rayner, S.; Xia, H.-M.; Gao, G.F.; Yuan, H.; Tang, Y.-P.; Luo, M.-H. Proteomic Analysis of Zika Virus Infected Primary Human Fetal Neural Progenitors Suggests a Role for Doublecortin in the Pathological Consequences of Infection in the Cortex. Front. Microbiol. 2018, 9, 1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcez, P.P.; Nascimento, J.; De Vasconcelos, J.M.; Da Costa, R.M.; DelVecchio, R.; Trindade, P.; Loiola, E.; Higa, L.M.; Cassoli, J.S.; Vitória, G.; et al. Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep. 2017, 7, 40780. [Google Scholar] [CrossRef]
- Richardson, R.B.; Ohlson, M.B.; Eitson, J.L.; Kumar, A.; McDougal, M.B.; Boys, I.N.; Mar, K.B.; De La Cruz-Rivera, P.C.; Douglas, C.; Konopka, G.; et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018, 3, 1214–1223. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Persson, B.D.; Nord, S.; Lindqvist, R.; Danskog, K.; Överby, A.K.; Kohl, A.; Willison, H.J.; Lenman, A.; Arnberg, N. BAF45b Is Required for Efficient Zika Virus Infection of HAP1 Cells. Viruses 2021, 13, 2007. https://doi.org/10.3390/v13102007
Persson BD, Nord S, Lindqvist R, Danskog K, Överby AK, Kohl A, Willison HJ, Lenman A, Arnberg N. BAF45b Is Required for Efficient Zika Virus Infection of HAP1 Cells. Viruses. 2021; 13(10):2007. https://doi.org/10.3390/v13102007
Chicago/Turabian StylePersson, B. David, Stefan Nord, Richard Lindqvist, Katarina Danskog, Anna K. Överby, Alain Kohl, Hugh J. Willison, Annasara Lenman, and Niklas Arnberg. 2021. "BAF45b Is Required for Efficient Zika Virus Infection of HAP1 Cells" Viruses 13, no. 10: 2007. https://doi.org/10.3390/v13102007





