Facilitating Antiviral Drug Discovery Using Genetic and Evolutionary Knowledge
Abstract
:1. Introduction
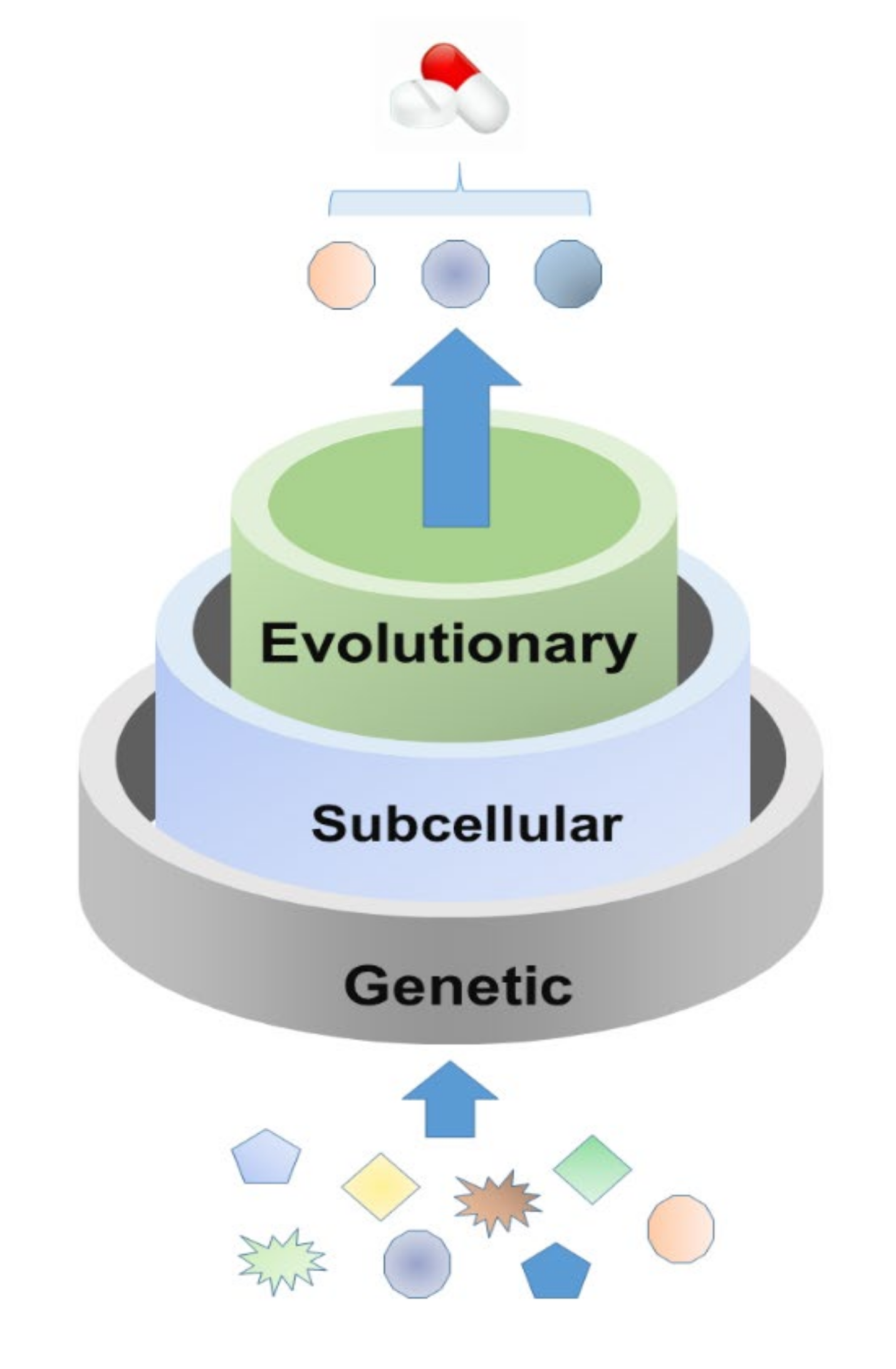
2. Feature Analysis for Host Targets of Approved Antiviral Drugs
2.1. Genetic Features
2.2. Subcellular Location Features
2.3. Evolutionary Features
3. Rational Screening of Potential Antiviral Host Targets
3.1. Target Screening by Summarized Features
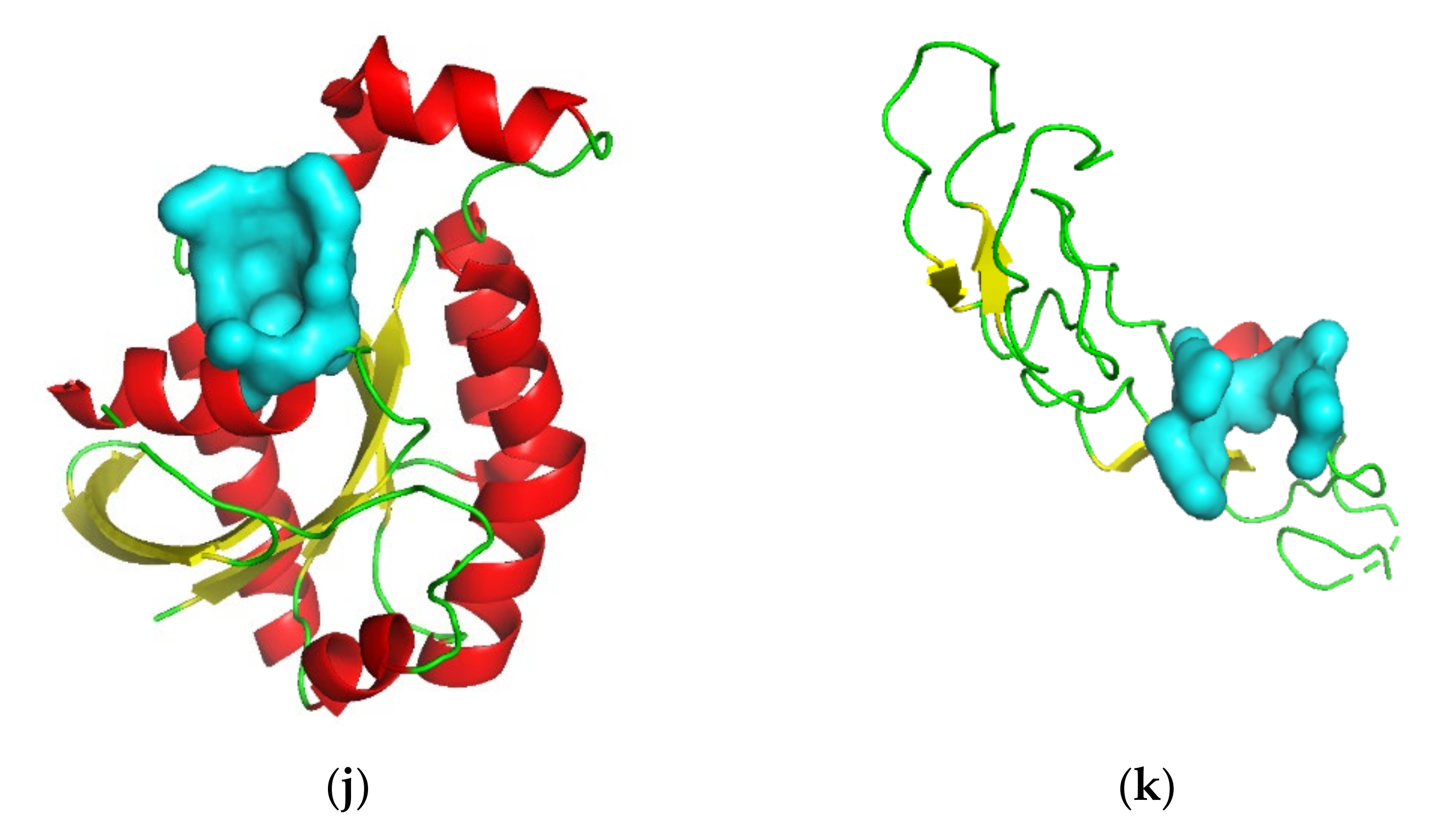
3.2. Target Screening by Protein Structures
4. Antiviral Drug Discovery Based on Screened Host Targets
4.1. Drug Repositioning
4.2. De Novo Drug Discovery
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Woolhouse, M.; Scott, F.; Hudson, Z.; Howey, R.; Chase-Topping, M. Human Viruses: Discovery and Emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2864–2871. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Hasan, S.; Ahmad, S.A.; Masood, R.; Saeed, S. Ebola Virus: A Global Public Health Menace: A Narrative Review. J. Fam. Med. Prim. Care 2019, 8, 2189–2201. [Google Scholar] [CrossRef]
- Marshall, A.H.; Rachlis, A.; Chen, J. Severe Acute Respiratory Syndrome: Responses of the Healthcare System to a Global Epidemic. Curr. Opin. Otolaryngol. Head Neck Surg. 2005, 13, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.J. Epstein–Barr Virus and Cancer. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zou, Z.; Dong, B.; Zhang, W.; Chen, T.; Cui, W.; Ma, Y. Secular Trends in HIV/AIDS Mortality in China from 1990 to 2016: Gender Disparities. PLoS ONE 2019, 14, e0219689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Singh, A.K. Plague Vaccine: Recent Progress and Prospects. npj Vaccines 2019, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maartens, G.; Celum, C.; Lewin, S.R. HIV Infection: Epidemiology, Pathogenesis, Treatment, and Prevention. Lancet 2014, 384, 258–271. [Google Scholar] [CrossRef]
- Ji, X.; Li, Z. Medicinal Chemistry Strategies toward Host Targeting Antiviral Agents. Med. Res. Rev. 2020, 40, 1519–1557. [Google Scholar] [CrossRef]
- Pillet, S.; Pozzetto, B.; Roblin, X. Cytomegalovirus and Ulcerative Colitis: Place of Antiviral Therapy. World J. Gastroenterol. 2016, 22, 2030–2045. [Google Scholar] [CrossRef]
- Mareri, A.; Lasorella, S.; Iapadre, G.; Maresca, M.; Tambucci, R.; Nigro, G. Anti-Viral Therapy for Congenital Cytomegalovirus Infection: Pharmacokinetics, Efficacy and Side Effects. J. Matern. Fetal. Neonatal. Med. 2016, 29, 1657–1664. [Google Scholar] [CrossRef]
- Jackson, W.E.; Everson, G.T. Sofosbuvir and Velpatasvir for the Treatment of Hepatitis C. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 501–505. [Google Scholar] [CrossRef]
- Schaefer, E.A.K.; Chung, R.T. Anti-Hepatitis C Virus Drugs in Development. Gastroenterology 2012, 142, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Selective Anti-Herpesvirus Agents. Antivir. Chem. Chemother. 2013, 23, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, K. Antiviral Drugs Against Alphaherpesvirus. Adv. Exp. Med. Biol. 2018, 1045, 103–122. [Google Scholar] [CrossRef]
- Amarelle, L.; Lecuona, E.; Sznajder, J.I. Anti-Influenza Treatment: Drugs Currently Used and Under Development. Arch. Bronconeumol. 2017, 53, 19–26. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Li, G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef] [Green Version]
- Menéndez-Arias, L.; Richman, D.D. Editorial Overview: Antivirals and Resistance: Advances and Challenges Ahead. Curr. Opin. Virol. 2014, 8, iv–vii. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi Pour, P.; Fakhri, S.; Asgary, S.; Farzaei, M.H.; Echeverría, J. The Signaling Pathways, and Therapeutic Targets of Antiviral Agents: Focusing on the Antiviral Approaches and Clinical Perspectives of Anthocyanins in the Management of Viral Diseases. Front. Pharmacol. 2019, 10, 1207. [Google Scholar] [CrossRef] [Green Version]
- Brodniewicz, T.; Grynkiewicz, G. Preclinical Drug Development. Acta Pol. Pharm. 2010, 67, 578–585. [Google Scholar] [PubMed]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the Decline in Pharmaceutical R&D Efficiency. Nat. Rev. Drug. Discov. 2012, 11, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Tufts Study Pegs Drug Development, Approval Cost at $2.6B. GEN—Genetic Engineering Biotechnology News, 18 November 2014.
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical Development Success Rates for Investigational Drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Munos, B. Lessons from 60 Years of Pharmaceutical Innovation. Nat. Rev. Drug. Discov. 2009, 8, 959–968. [Google Scholar] [CrossRef]
- Vandamme, D.; Minke, B.A.; Fitzmaurice, W.; Kholodenko, B.N.; Kolch, W. Systems Biology-Embedded Target Validation: Improving Efficacy in Drug Discovery. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 1–11. [Google Scholar] [CrossRef]
- Quan, Y.; Luo, Z.-H.; Yang, Q.-Y.; Li, J.; Zhu, Q.; Liu, Y.-M.; Cui, Z.-J.; Qin, X.; Lv, B.-M.; Xu, Y.-H.; et al. Systems Chemical Genetics-Based Drug Discovery: Prioritizing Agents Targeting Multiple/Reliable Disease-Associated Genes as Drug Candidates. Front. Genet. 2019, 10, 474. [Google Scholar] [CrossRef] [Green Version]
- Nikonov, O.S.; Chernykh, E.S.; Garber, M.B.; Nikonova, E.Y. Enteroviruses: Classification, Diseases They Cause, and Approaches to Development of Antiviral Drugs. Biochem. Mosc. 2017, 82, 1615–1631. [Google Scholar] [CrossRef]
- Prussia, A.; Thepchatri, P.; Snyder, J.P.; Plemper, R.K. Systematic Approaches towards the Development of Host-Directed Antiviral Therapeutics. Int. J. Mol. Sci. 2011, 12, 4027–4052. [Google Scholar] [CrossRef] [Green Version]
- Münk, C.; Sommer, A.F.R.; König, R. Systems-Biology Approaches to Discover Anti-Viral Effectors of the Human Innate Immune Response. Viruses 2011, 3, 1112–1130. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.J.; Hill, A.V.S. Human Genetic Susceptibility to Infectious Disease. Nat. Rev. Genet. 2012, 13, 175–188. [Google Scholar] [CrossRef]
- Gelbart, M.; Harari, S.; Ben-Ari, Y.; Kustin, T.; Wolf, D.; Mandelboim, M.; Mor, O.; Pennings, P.S.; Stern, A. Drivers of Within-Host Genetic Diversity in Acute Infections of Viruses. PLoS Pathog. 2020, 16, e1009029. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Ranjan, K.; Brar, B.; Shah, I.; Lalmbe, U.; Manimegalai, J.; Vashisht, B.; Gaury, M.; Kumar, P.; Khurana, S.K.; et al. Virus-Host Interactions: New Insights and Advances in Drug Development Against Viral Pathogens. Curr. Drug Metab. 2017, 18, 942–970. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.; Hou, W.; Dakshanamurthy, S.; Tang, Q. Host Targeted Antiviral (HTA): Functional Inhibitor Compounds of Scaffold Protein RACK1 Inhibit Herpes Simplex Virus Proliferation. Oncotarget 2019, 10, 3209–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyrrell, B.E.; Sayce, A.C.; Warfield, K.L.; Miller, J.L.; Zitzmann, N. Iminosugars: Promising Therapeutics for Influenza Infection. Crit. Rev. Microbiol. 2017, 43, 521–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plummer, E.; Buck, M.D.; Sanchez, M.; Greenbaum, J.A.; Turner, J.; Grewal, R.; Klose, B.; Sampath, A.; Warfield, K.L.; Peters, B.; et al. Dengue Virus Evolution under a Host-Targeted Antiviral. J. Virol. 2015, 89, 5592–5601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villalón-Letelier, F.; Brooks, A.G.; Saunders, P.M.; Londrigan, S.L.; Reading, P.C. Host Cell Restriction Factors That Limit Influenza A Infection. Viruses 2017, 9, 376. [Google Scholar] [CrossRef] [Green Version]
- Glennon, E.K.K.; Dankwa, S.; Smith, J.D.; Kaushansky, A. Opportunities for Host-Targeted Therapies for Malaria. Trends Parasitol. 2018, 34, 843–860. [Google Scholar] [CrossRef]
- Varikuti, S.; Jha, B.K.; Volpedo, G.; Ryan, N.M.; Halsey, G.; Hamza, O.M.; McGwire, B.S.; Satoskar, A.R. Host-Directed Drug Therapies for Neglected Tropical Diseases Caused by Protozoan Parasites. Front. Microbiol. 2018, 9, 2655. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Hansen, E.; Cunningham, J.J.; Rego, E.H.; Turner, P.E.; Weitz, J.S.; Hochberg, M.E. Fighting Microbial Pathogens by Integrating Host Ecosystem Interactions and Evolution. Bioessays 2021, 43, e2000272. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Fu, L.-Y.; Zhang, H.-Y. Can Medical Genetics and Evolutionary Biology Inspire Drug Target Identification? Trends Mol. Med. 2012, 18, 69–71. [Google Scholar] [CrossRef]
- Kamb, A.; Harper, S.; Stefansson, K. Human Genetics as a Foundation for Innovative Drug Development. Nat. Biotechnol. 2013, 31, 975–978. [Google Scholar] [CrossRef]
- Quan, Y.; Wang, Z.Y.; Chu, X.Y.; Zhang, H.Y. Evolutionary and Genetic Features of Drug Targets. Med. Res. Rev. 2018, 38, 1536–1549. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; García-Sastre, A. Antiviral Innate Immunity through the Lens of Systems Biology. Virus Res. 2016, 218, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenney, A.D.; Dowdle, J.A.; Bozzacco, L.; McMichael, T.M.; St. Gelais, C.; Panfil, A.R.; Sun, Y.; Schlesinger, L.S.; Anderson, M.Z.; Green, P.L.; et al. Human Genetic Determinants of Viral Diseases. Annu. Rev. Genet. 2017, 51, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Klebanov, N. Genetic Predisposition to Infectious Disease. Cureus 2018, 10, e3210. [Google Scholar] [CrossRef] [Green Version]
- Yudin, N.S.; Barkhash, A.V.; Maksimov, V.N.; Ignatieva, E.V.; Romaschenko, A.G. Human Genetic Predisposition to Diseases Caused by Viruses from Flaviviridae Family. Mol. Biol. 2018, 52, 190–209. [Google Scholar] [CrossRef]
- Akcay, I.M.; Katrinli, S.; Ozdil, K.; Doganay, G.D.; Doganay, L. Host Genetic Factors Affecting Hepatitis B Infection Outcomes: Insights from Genome-Wide Association Studies. World J. Gastroenterol. 2018, 24, 3347–3360. [Google Scholar] [CrossRef]
- Verma, R.; Sharma, P.C. Next Generation Sequencing-Based Emerging Trends in Molecular Biology of Gastric Cancer. Am. J. Cancer Res. 2018, 8, 207–225. [Google Scholar]
- Ma, B.B.Y.; Hui, E.P.; Chan, A.T.C. Investigational Drugs for Nasopharyngeal Carcinoma. Expert Opin. Investig. Drugs 2017, 26, 677–685. [Google Scholar] [CrossRef]
- Hashemi, S.M.A.; Thijssen, M.; Hosseini, S.Y.; Tabarraei, A.; Pourkarim, M.R.; Sarvari, J. Human Gene Polymorphisms and Their Possible Impact on the Clinical Outcome of SARS-CoV-2 Infection. Arch. Virol. 2021, 166, 1–20. [Google Scholar] [CrossRef]
- Guedj, J.; Yu, J.; Levi, M.; Li, B.; Kern, S.; Naoumov, N.V.; Perelson, A.S. Modeling Viral Kinetics and Treatment Outcome during Alisporivir Interferon-Free Treatment in Hepatitis C Virus Genotype 2 and 3 Patients. Hepatology 2014, 59, 1706–1714. [Google Scholar] [CrossRef]
- Nissen, S.K.; Christiansen, M.; Helleberg, M.; Kjær, K.; Jørgensen, S.E.; Gerstoft, J.; Katzenstein, T.L.; Benfield, T.; Kronborg, G.; Larsen, C.S.; et al. Whole Exome Sequencing of HIV-1 Long-Term Non-Progressors Identifies Rare Variants in Genes Encoding Innate Immune Sensors and Signaling Molecules. Sci. Rep. 2018, 8, 15253. [Google Scholar] [CrossRef] [PubMed]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Brest, P.; Mograbi, B.; Hofman, P.; Milano, G. Using Genetics To Dissect SARS-CoV-2 Infection. Trends Genet. 2021, 37, 203–204. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, A.H.; Snijder, E.J.; Kikkert, M.; van Hemert, M.J. Host Factors in Coronavirus Replication. Curr. Top. Microbiol. Immunol. 2018, 419, 1–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Yang, J.; Feng, B.; Lu, W.; Zhao, C.; Li, L. Mendelian Randomization Analysis Identified Genes Pleiotropically Associated with the Risk and Prognosis of COVID-19. J. Infect. 2021, 82, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public Archive of Relationships among Sequence Variation and Human Phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.Org: Online Mendelian Inheritance in Man (OMIM®), an Online Catalog of Human Genes and Genetic Disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [Green Version]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a Comprehensive Repository of Inherited Mutation Data for Medical Research, Genetic Diagnosis and next-Generation Sequencing Studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavan, S.; Rommel, K.; Mateo Marquina, M.E.; Höhn, S.; Lanneau, V.; Rath, A. Clinical Practice Guidelines for Rare Diseases: The Orphanet Database. PLoS ONE 2017, 12, e0170365. [Google Scholar] [CrossRef] [Green Version]
- Li, M.J.; Liu, Z.; Wang, P.; Wong, M.P.; Nelson, M.R.; Kocher, J.-P.A.; Yeager, M.; Sham, P.C.; Chanock, S.J.; Xia, Z.; et al. GWASdb v2: An Update Database for Human Genetic Variants Identified by Genome-Wide Association Studies. Nucleic Acids Res. 2016, 44, D869–D876. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Tian, W. Explaining the Disease Phenotype of Intergenic SNP through Predicted Long Range Regulation. Nucleic Acids Res. 2016, 44, 8641–8654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, K.G.; Barnes, K.C.; Bright, T.J.; Wang, S.A. The Genetic Association Database. Nat. Genet. 2004, 36, 431–432. [Google Scholar] [CrossRef] [Green Version]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A Discovery Platform for the Dynamical Exploration of Human Diseases and Their Genes. Database 2015, 2015, bav028. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, M.D.; Fradet-Turcotte, A. Virus DNA Replication and the Host DNA Damage Response. Annu. Rev. Virol. 2018, 5, 141–164. [Google Scholar] [CrossRef]
- Boulant, S.; Stanifer, M.; Lozach, P.-Y. Dynamics of Virus-Receptor Interactions in Virus Binding, Signaling, and Endocytosis. Viruses 2015, 7, 2794–2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maginnis, M.S. Virus—Receptor Interactions: The Key to Cellular Invasion. J. Mol. Biol. 2018, 430, 2590–2611. [Google Scholar] [CrossRef]
- Rajendran, L.; Knölker, H.-J.; Simons, K. Subcellular Targeting Strategies for Drug Design and Delivery. Nat. Rev. Drug. Discov. 2010, 9, 29–42. [Google Scholar] [CrossRef]
- Clapham, P.R.; McKnight, Á. HIV-1 Receptors and Cell Tropism. Br. Med. Bull. 2001, 58, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Sundaravaradan, V.; Das, S.R.; Ramakrishnan, R.; Sehgal, S.; Gopalan, S.; Ahmad, N.; Jameel, S. Role of HIV-1 Subtype C Envelope V3 to V5 Regions in Viral Entry, Coreceptor Utilization and Replication Efficiency in Primary T-Lymphocytes and Monocyte-Derived Macrophages. Virol. J. 2007, 4, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.B.; Giesler, K.E.; Tahirovic, Y.A.; Truax, V.M.; Liotta, D.C.; Wilson, L.J. CCR5 Receptor Antagonists in Preclinical to Phase II Clinical Development for Treatment of HIV. Expert Opin. Investig. Drugs 2016, 25, 1377–1392. [Google Scholar] [CrossRef] [Green Version]
- Lenz, J.C.C.; Rockstroh, J.K. Vicriviroc, a New CC-Chemokine Receptor 5 Inhibitor for Treatment of HIV: Properties, Promises and Challenges. Expert Opin. Drug Metab. Toxicol. 2010, 6, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [Green Version]
- Gorter, F.A.; Hall, A.R.; Buckling, A.; Scanlan, P.D. Parasite Host Range and the Evolution of Host Resistance. J. Evol. Biol. 2015, 28, 1119–1130. [Google Scholar] [CrossRef]
- Dallas, T.; Huang, S.; Nunn, C.; Park, A.W.; Drake, J.M. Estimating Parasite Host Range. Proc. Biol. Sci. 2017, 284, 20171250. [Google Scholar] [CrossRef] [Green Version]
- Paterson, S.; Vogwill, T.; Buckling, A.; Benmayor, R.; Spiers, A.J.; Thomson, N.R.; Quail, M.; Smith, F.; Walker, D.; Libberton, B.; et al. Antagonistic Coevolution Accelerates Molecular Evolution. Nature 2010, 464, 275–278. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V. Viruses and Mobile Elements as Drivers of Evolutionary Transitions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150442. [Google Scholar] [CrossRef]
- Voskarides, K.; Christaki, E.; Nikolopoulos, G.K. Influenza Virus—Host Co-Evolution. A Predator-Prey Relationship? Front. Immunol. 2018, 9, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.E.; Busse, D.C.; Binter, S.; Weston, S.; Diaz Soria, C.; Laksono, B.M.; Clare, S.; van Nieuwkoop, S.; van den Hoogen, B.G.; Clement, M.; et al. Interferon-Induced Transmembrane Protein 1 Restricts Replication of Viruses That Enter Cells via the Plasma Membrane. J. Virol. 2019, 93, e02003-18. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Franco, L.M.; Atmar, R.L.; Quarles, J.M.; Arden, N.; Bucasas, K.L.; Wells, J.M.; Niño, D.; Wang, X.; Zapata, G.E.; et al. Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections—A Prospective Cohort Study. PLoS Pathog. 2015, 11, e1004869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, J.J.; Levin, B.R.; Molineux, I.J. Promises and Pitfalls of In Vivo Evolution to Improve Phage Therapy. Viruses 2019, 11, 1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurney, J.; Brown, S.P.; Kaltz, O.; Hochberg, M.E. Steering Phages to Combat Bacterial Pathogens. Trends Microbiol. 2020, 28, 85–94. [Google Scholar] [CrossRef]
- Phillips, A.M.; Gonzalez, L.O.; Nekongo, E.E.; Ponomarenko, A.I.; McHugh, S.M.; Butty, V.L.; Levine, S.S.; Lin, Y.-S.; Mirny, L.A.; Shoulders, M.D. Host Proteostasis Modulates Influenza Evolution. eLife 2017, 6, e28652. [Google Scholar] [CrossRef] [Green Version]
- Domazet-Lošo, T.; Tautz, D. Phylostratigraphic Tracking of Cancer Genes Suggests a Link to the Emergence of Multicellularity in Metazoa. BMC Biol. 2010, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Diamond, M.S.; Farzan, M. The Broad-Spectrum Antiviral Functions of IFIT and IFITM Proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef]
- Liebeskind, B.J.; McWhite, C.D.; Marcotte, E.M. Towards Consensus Gene Ages. Genome Biol. Evol. 2016, 8, 1812–1823. [Google Scholar] [CrossRef]
- Domazet-Lošo, T.; Carvunis, A.-R.; Albà, M.M.; Šestak, M.S.; Bakaric, R.; Neme, R.; Tautz, D. No Evidence for Phylostratigraphic Bias Impacting Inferences on Patterns of Gene Emergence and Evolution. Mol. Biol. Evol. 2017, 34, 843–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnhammer, E.L.L.; Gabaldón, T.; Sousa da Silva, A.W.; Martin, M.; Robinson-Rechavi, M.; Boeckmann, B.; Thomas, P.D.; Dessimoz, C. Quest for Orthologs consortium Big Data and Other Challenges in the Quest for Orthologs. Bioinformatics 2014, 30, 2993–2998. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B.; Marcet-Houben, M.; Rees, J.A.; Forslund, K.; Huerta-Cepas, J.; Muffato, M.; Yilmaz, P.; Xenarios, I.; Bork, P.; Lewis, S.E.; et al. Quest for Orthologs Entails Quest for Tree of Life: In Search of the Gene Stream. Genome Biol. Evol. 2015, 7, 1988–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plenge, R.M.; Scolnick, E.M.; Altshuler, D. Validating Therapeutic Targets through Human Genetics. Nat. Rev. Drug. Discov. 2013, 12, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Cong, F.; Cheung, A.K.; Huang, S.-M.A. Chemical Genetics-Based Target Identification in Drug Discovery. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 57–78. [Google Scholar] [CrossRef]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotto, K.C.; Wagner, A.H.; Feng, Y.-Y.; Kiwala, S.; Coffman, A.C.; Spies, G.; Wollam, A.; Spies, N.C.; Griffith, O.L.; Griffith, M. DGIdb 3.0: A Redesign and Expansion of the Drug-Gene Interaction Database. Nucleic Acids Res. 2018, 46, D1068–D1073. [Google Scholar] [CrossRef] [Green Version]
- Betz, U.A.K. How Many Genomics Targets Can a Portfolio Afford? Drug Discov. Today 2005, 10, 1057–1063. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Groom, C.R. The Druggable Genome. Nat. Rev. Drug. Discov. 2002, 1, 727–730. [Google Scholar] [CrossRef]
- Betz, U.A.; Farquhar, R.; Ziegelbauer, K. Genomics: Success or Failure to Deliver Drug Targets? Curr. Opin. Chem. Biol. 2005, 9, 387–391. [Google Scholar] [CrossRef]
- Borrel, A.; Regad, L.; Xhaard, H.; Petitjean, M.; Camproux, A.-C. PockDrug: A Model for Predicting Pocket Druggability That Overcomes Pocket Estimation Uncertainties. J. Chem. Inf. Model. 2015, 55, 882–895. [Google Scholar] [CrossRef]
- Hajduk, P.J.; Huth, J.R.; Fesik, S.W. Druggability Indices for Protein Targets Derived from NMR-Based Screening Data. J. Med. Chem. 2005, 48, 2518–2525. [Google Scholar] [CrossRef] [PubMed]
- Kana, O.; Brylinski, M. Elucidating the Druggability of the Human Proteome with EFindSite. J. Comput. Aided Mol. Des. 2019, 33, 509–519. [Google Scholar] [CrossRef]
- Fernández, A. Deep Learning to Therapeutically Target Unreported Complexes. Trends Pharmacol. Sci. 2019, 40, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological Macromolecular Structures Enabling Research and Education in Fundamental Biology, Biomedicine, Biotechnology and Energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Y.; Zhang, H.-Y. Rational Drug Repositioning by Medical Genetics. Nat. Biotechnol. 2013, 31, 1080–1082. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yao, Y.; Cheng, X.; Chen, Q.; Zhao, W.; Ma, S.; Li, Z.; Zhou, H.; Li, W.; Fei, T. A Computational Framework of Host-Based Drug Repositioning for Broad-Spectrum Antivirals against RNA Viruses. iScience 2021, 24, 102148. [Google Scholar] [CrossRef]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The Family of the Interleukin-1 Receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, N.; Ji, H.; Huang, Y.; Hu, H.; Li, J.; Mi, S.; Duan, S.; Chen, X. A Male-Specific Association between AGTR1 Hypermethylation and Coronary Heart Disease. Bosn. J. Basic Med. Sci. 2020, 20, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Al-Salama, Z.T.; Frampton, J.E. Glycopyrronium/Formoterol: A Review in COPD. Drugs 2019, 79, 1455–1466. [Google Scholar] [CrossRef]
- Engering, A.; Geijtenbeek, T.B.H.; van Vliet, S.J.; Wijers, M.; van Liempt, E.; Demaurex, N.; Lanzavecchia, A.; Fransen, J.; Figdor, C.G.; Piguet, V.; et al. The Dendritic Cell-Specific Adhesion Receptor DC-SIGN Internalizes Antigen for Presentation to T Cells. J. Immunol. 2002, 168, 2118–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, G.; Simmons, G.; Pöhlmann, S.; Baribaud, F.; Ni, H.; Leslie, G.J.; Haggarty, B.S.; Bates, P.; Weissman, D.; Hoxie, J.A.; et al. Differential N-Linked Glycosylation of Human Immunodeficiency Virus and Ebola Virus Envelope Glycoproteins Modulates Interactions with DC-SIGN and DC-SIGNR. J. Virol. 2003, 77, 1337–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.A.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.L.; et al. DC-SIGN (CD209) Mediates Dengue Virus Infection of Human Dendritic Cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef] [Green Version]
- Glinka, A.; Dolde, C.; Kirsch, N.; Huang, Y.-L.; Kazanskaya, O.; Ingelfinger, D.; Boutros, M.; Cruciat, C.-M.; Niehrs, C. LGR4 and LGR5 Are R-Spondin Receptors Mediating Wnt/β-Catenin and Wnt/PCP Signalling. EMBO Rep. 2011, 12, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Becker, L.; Huang, Q.; Mashimo, H. Immunostaining of Lgr5, an Intestinal Stem Cell Marker, in Normal and Premalignant Human Gastrointestinal Tissue. Sci. World J. 2008, 8, 1168–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-Based Propagation of Functional Annotations within the Gene Ontology Consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Cascino, I.; Fiucci, G.; Papoff, G.; Ruberti, G. Three Functional Soluble Forms of the Human Apoptosis-Inducing Fas Molecule Are Produced by Alternative Splicing. J. Immunol. 1995, 154, 2706–2713. [Google Scholar]
- Cao, P.; Zhang, W.; Gui, W.; Dong, Y.; Jiang, T.; Gong, Y. Structural Insights into the Mechanism of Calmodulin Binding to Death Receptors. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1604–1613. [Google Scholar] [CrossRef]
- Oehm, A.; Behrmann, I.; Falk, W.; Pawlita, M.; Maier, G.; Klas, C.; Li-Weber, M.; Richards, S.; Dhein, J.; Trauth, B.C. Purification and Molecular Cloning of the APO-1 Cell Surface Antigen, a Member of the Tumor Necrosis Factor/Nerve Growth Factor Receptor Superfamily. Sequence Identity with the Fas Antigen. J. Biol. Chem. 1992, 267, 10709–10715. [Google Scholar] [CrossRef]
- Fang, F.; Lue, L.-F.; Yan, S.; Xu, H.; Luddy, J.S.; Chen, D.; Walker, D.G.; Stern, D.M.; Yan, S.; Schmidt, A.M.; et al. RAGE-Dependent Signaling in Microglia Contributes to Neuroinflammation, Aβ Accumulation, and Impaired Learning/Memory in a Mouse Model of Alzheimer’s Disease. FASEB J. 2010, 24, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Rai, V.; Singer, D.; Chabierski, S.; Xie, J.; Reverdatto, S.; Burz, D.S.; Schmidt, A.M.; Hoffmann, R.; Shekhtman, A. Advanced Glycation End Product Recognition by the Receptor for AGEs. Structure 2011, 19, 722–732. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Shi, Z.; Baumgart, T. Mutations in BIN1 Associated with Centronuclear Myopathy Disrupt Membrane Remodeling by Affecting Protein Density and Oligomerization. PLoS ONE 2014, 9, e93060. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, T.; Ebinuma, I.; Morohashi, Y.; Hori, Y.; Young Chang, M.; Hattori, H.; Maehara, T.; Yokoshima, S.; Fukuyama, T.; Tsuji, S.; et al. BIN1 Regulates BACE1 Intracellular Trafficking and Amyloid-β Production. Hum. Mol. Genet. 2016, 25, 2948–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lingel, A.; Weiss, T.M.; Niebuhr, M.; Pan, B.; Appleton, B.A.; Wiesmann, C.; Bazan, J.F.; Fairbrother, W.J. Structure of IL-33 and Its Interaction with the ST2 and IL-1RAcP Receptors-Insight into Heterotrimeric IL-1 Signaling Complexes. Structure 2009, 17, 1398–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Hammel, M.; He, Y.; Tainer, J.A.; Jeng, U.-S.; Zhang, L.; Wang, S.; Wang, X. Structural Insights into the Interaction of IL-33 with Its Receptors. Proc. Natl. Acad. Sci. USA 2013, 110, 14918–14923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, R.G.; Abo, A.; Steven Martin, G. A Human Homolog of the C. Elegans Polarity Determinant Par-6 Links Rac and Cdc42 to PKCzeta Signaling and Cell Transformation. Curr. Biol. 2000, 10, 697–707. [Google Scholar] [CrossRef] [Green Version]
- Drenan, R.M.; Doupnik, C.A.; Boyle, M.P.; Muglia, L.J.; Huettner, J.E.; Linder, M.E.; Blumer, K.J. Palmitoylation Regulates Plasma Membrane-Nuclear Shuttling of R7BP, a Novel Membrane Anchor for the RGS7 Family. J. Cell Biol. 2005, 169, 623–633. [Google Scholar] [CrossRef]
- Hunt, R.A.; Edris, W.; Chanda, P.K.; Nieuwenhuijsen, B.; Young, K.H. Snapin Interacts with the N-Terminus of Regulator of G Protein Signaling 7. Biochem. Biophys. Res. Commun. 2003, 303, 594–599. [Google Scholar] [CrossRef]
- Tao, W.; Pennica, D.; Xu, L.; Kalejta, R.F.; Levine, A.J. Wrch-1, a Novel Member of the Rho Gene Family That Is Regulated by Wnt-1. Genes Dev. 2001, 15, 1796–1807. [Google Scholar] [CrossRef] [Green Version]
- Pitti, R.M.; Marsters, S.A.; Ruppert, S.; Donahue, C.J.; Moore, A.; Ashkenazi, A. Induction of Apoptosis by Apo-2 Ligand, a New Member of the Tumor Necrosis Factor Cytokine Family. J. Biol. Chem. 1996, 271, 12687–12690. [Google Scholar] [CrossRef] [Green Version]
- Ramamurthy, V.; Yamniuk, A.P.; Lawrence, E.J.; Yong, W.; Schneeweis, L.A.; Cheng, L.; Murdock, M.; Corbett, M.J.; Doyle, M.L.; Sheriff, S. The Structure of the Death Receptor 4-TNF-Related Apoptosis-Inducing Ligand (DR4-TRAIL) Complex. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 1273–1281. [Google Scholar] [CrossRef] [Green Version]
- Doud, M.B.; Lee, J.M.; Bloom, J.D. How Single Mutations Affect Viral Escape from Broad and Narrow Antibodies to H1 Influenza Hemagglutinin. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, M.C.; Shirey, K.A.; Pletneva, L.M.; Boukhvalova, M.S.; Garzino-Demo, A.; Vogel, S.N.; Blanco, J.C. Novel Drugs Targeting Toll-like Receptors for Antiviral Therapy. Future Virol. 2014, 9, 811–829. [Google Scholar] [CrossRef] [Green Version]
- Brelot, A.; Chakrabarti, L.A. CCR5 Revisited: How Mechanisms of HIV Entry Govern AIDS Pathogenesis. Available online: https://pubmed.ncbi.nlm.nih.gov/29932942/?from_term=CCR5+&from_pos=2 (accessed on 18 May 2020).
- Grande, F.; Occhiuzzi, M.A.; Rizzuti, B.; Ioele, G.; De Luca, M.; Tucci, P.; Svicher, V.; Aquaro, S.; Garofalo, A. CCR5/CXCR4 Dual Antagonism for the Improvement of HIV Infection Therapy. Molecules 2019, 24, 550. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Sharma, S.; Kumar, R.; Tripathi, B.N.; Barua, S.; Ly, H.; Rouse, B.T. Host-Directed Antiviral Therapy. Clin. Microbiol. Rev. 2020, 33, e00168-19. [Google Scholar] [CrossRef]
- McHutchison, J.G.; Manns, M.P.; Muir, A.J.; Terrault, N.A.; Jacobson, I.M.; Afdhal, N.H.; Heathcote, E.J.; Zeuzem, S.; Reesink, H.W.; Garg, J.; et al. Telaprevir for Previously Treated Chronic HCV Infection. N. Engl. J. Med. 2010, 362, 1292–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imran, M.; Manzoor, S.; Khattak, N.M.; Khalid, M.; Ahmed, Q.L.; Parvaiz, F.; Tariq, M.; Ashraf, J.; Ashraf, W.; Azam, S.; et al. Current and Future Therapies for Hepatitis C Virus Infection: From Viral Proteins to Host Targets. Arch. Virol. 2014, 159, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-Directed Therapies for Bacterial and Viral Infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef]
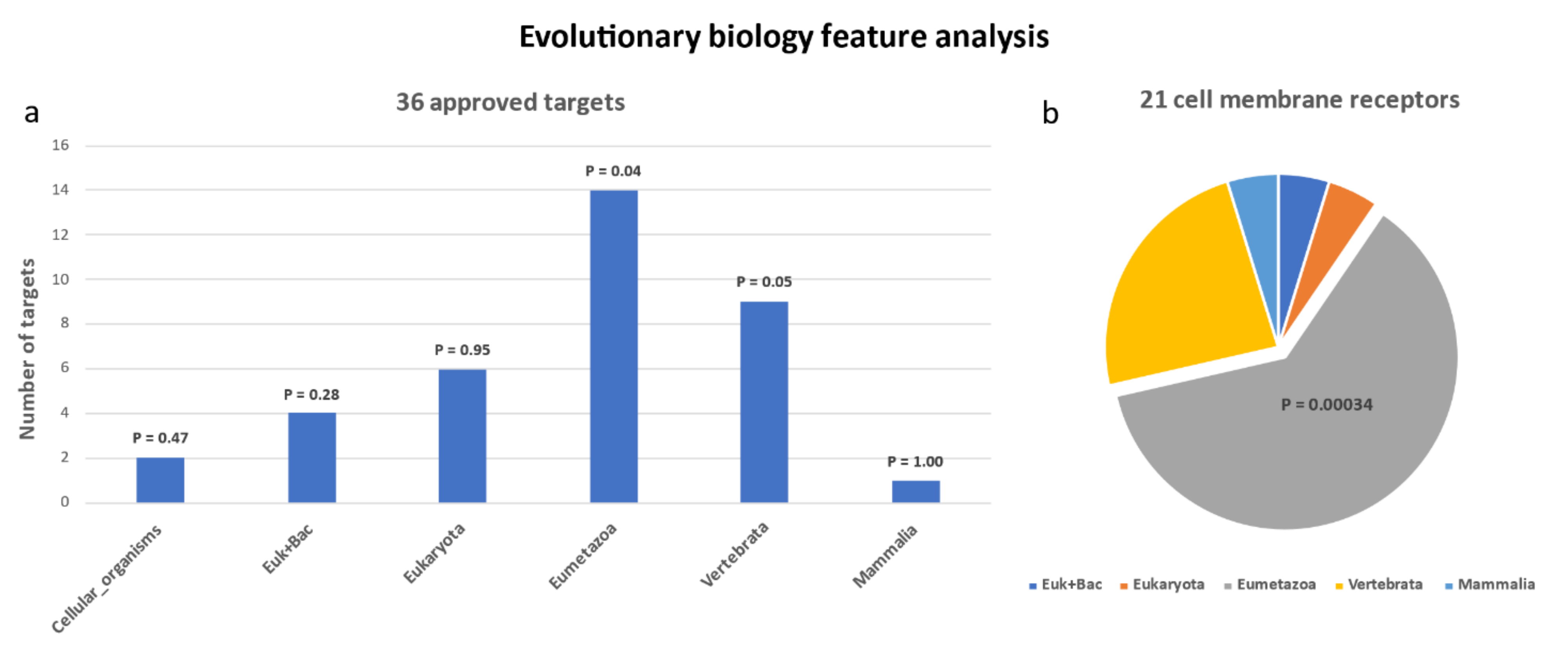

| Gene Symbol | Uniprot ID | Protein Name | SCG Virus Diseases Median Score | Subcellular Location | Origin | Associated Virus Diseases Name (Only List the Top 3) * |
|---|---|---|---|---|---|---|
| C1QA | P02745 | Complement C1q subcomponent subunit A | 1 | Secreted | Vertebrata | Herpes Simplex |
| C1QB | P02746 | Complement C1q subcomponent subunit B | 1 | Secreted | Vertebrata | Herpes Simplex; Carcinoma, Merkel Cell |
| C1QC | P02747 | Complement C1q subcomponent subunit C | 1 | Secreted | Vertebrata | Herpes Simplex; Adenoviridae Infections |
| C1R | P00736 | Complement C1r subcomponent, EC 3.4.21.41 | 3 | Secreted | Vertebrata | AIDS Dementia Complex |
| CCR5 | P51681 | C-C chemokine receptor type 5, C-C CKR-5, CC-CKR-5, CCR-5, CCR5 | 3 | Cell membrane | Eumetazoa | Hepatitis, Viral, Human; West Nile Fever; Hepatitis C |
| CD4 | P01730 | T-cell surface glycoprotein CD4 | 3 | Cell membrane | Vertebrata | Measles; Picornaviridae Infections; HIV Infections |
| CHRNA3 | P32297 | Neuronal acetylcholine receptor subunit alpha-3 | 3 | Cell membrane | Eumetazoa | AIDS Dementia Complex; Carcinoma, Merkel Cell; Influenza, Human |
| CHRNA4 | P43681 | Neuronal acetylcholine receptor subunit alpha-4 | 1 | Cell membrane | Eumetazoa | AIDS Dementia Complex; Influenza, Human; Picornaviridae Infections |
| CHRNA7 | P36544 | Neuronal acetylcholine receptor subunit alpha-7 | 3 | Cell membrane | Eumetazoa | AIDS Dementia Complex |
| CXCR4 | P61073 | C-X-C chemokine receptor type 4 | 3 | Cell membrane | Eumetazoa | Herpes Simplex; Papillomavirus Infections; Epstein-Barr Virus Infections |
| DRD2 | P14416 | D(2) dopamine receptor | 1 | Cell membrane | Eumetazoa | AIDS Dementia Complex; Hemorrhagic Fevers, Viral; Hepatitis |
| ENPP1 | P22413 | Ectonucleotide pyrphosphatase/phosphodiesterase family member 1 | 3 | Cell membrane | Eukaryota | West Nile Fever; Hepatitis; AIDS Dementia Complex |
| FCGR1A | P12314 | High affinity immunoglobulin gamma Fc receptor I | 1 | Cell membrane | Eumetazoa | Leukoencephalopathy, Progressive Multifocal; Hemorrhagic Fever, Ebola; Hemorrhagic Fevers, Viral |
| FCGR2A | P12318 | Low affinity immunoglobulin gamma Fc region receptor II-a | 2 | Cell membrane | Eumetazoa | Leukoencephalopathy, Progressive Multifocal; West Nile Fever; Picornaviridae Infections |
| FCGR2B | P31994 | Low affinity immunoglobulin gamma Fc region receptor II-b | 1 | Cell membrane | Eumetazoa | Picornaviridae Infections; Hemorrhagic Fevers, Viral; Hepatitis C |
| FCGR2C | P31995 | Low affinity immunoglobulin gamma Fc region receptor II-c | 1 | Cell membrane | Eumetazoa | HIV Infections; Herpes Simplex; Hepatitis C |
| FCGR3A | P08637 | Low affinity immunoglobulin gamma Fc region receptor III-A | 3 | Cell membrane | Eumetazoa | Herpesviridae Infections; Hepatitis, Viral, Human; Measles |
| FCGR3B | O75015 | Low affinity immunoglobulin gamma Fc region receptor III-B | 1 | Cell membrane | Eumetazoa | Influenza, Human; Picornaviridae Infections; West Nile Fever |
| GRIN3A | Q8TCU5 | Glutamate receptor ionotropic, NMDA 3A, GluN3A | 3 | Cell membrane | Vertebrata | AIDS Dementia Complex |
| HLA-B | P18465 | HLA class I histocompatibility antigen, B alpha chain | 3 | Cell membrane | Vertebrata | HIV Infections; Hepatitis; Picornaviridae Infections |
| IFNAR1 | P17181 | Interferon alpha/beta receptor 1 | 1 | Cell membrane | Vertebrata | Leukoencephalopathy, Progressive Multifocal; Hemorrhagic Fevers, Viral; West Nile Fever |
| IFNAR2 | P48551 | Interferon alpha/beta receptor 2 | 3 | Cell membrane | Vertebrata | Hepatitis; Picornaviridae Infections; Leukoencephalopathy, Progressive Multifocal |
| IMPDH1 | P20839 | Inosine-5’-monophosphate dehydrogenase 1 | —— | Cytoplasm | Cellular_ organisms | —— |
| IMPDH2 | P12268 | Inosine-5’-monophosphate dehydrogenase 2 | 1 | Nucleus | Cellular_ organisms | Sarcoma, Kaposi |
| NEU1 | Q99519 | N-acetyl-alpha-neuraminidase 1 | 2 | Cell membrane | Euk + Bac | Carcinoma, Merkel Cell; Leukoencephalopathy, Progressive Multifocal; Sarcoma, Kaposi |
| NEU2 | Q9Y3R4 | N-acetyl-alpha-neuraminidase 2 | —— | Cytoplasm | Euk + Bac | —— |
| NR1I2 | O75469 | Nuclear receptor subfamily 1 group I member 2 | 1 | Nucleus | Eumetazoa | Hepatitis; Measles; Paramyxoviridae Infections… |
| NT5C2 | P49902 | Cytosolic purine 5’-nucleotidase, EC 3.1.3.5 | 1 | Cytoplasm | Eukaryota | AIDS Dementia Complex; Cytomegalovirus Infections; Sarcoma, Kaposi |
| PNP | P00491 | Purine nucleoside phosphorylase, PNP, EC 2.4.2.1 | 1 | Cytoplasm | Euk + Bac | Cytomegalovirus Infections; AIDS Dementia Complex; Epstein-Barr Virus Infections |
| SLCO1B1 | Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | 3 | Cell membrane | Mammalia | Fatigue Syndrome, Chronic; Picornaviridae Infections; HIV Infections |
| SLCO2B1 | O94956 | Solute carrier organic anion transporter family member 2B1 | —— | Cell membrane | Eumetazoa | —— |
| TERT | O14746 | Telomerase reverse transcriptase, EC 2.7.7.49 | 1 | Nucleolus | Eukaryota | Carcinoma, Merkel Cell; Leukoencephalopathy, Progressive Multifocal; Influenza, Human |
| TOP2A | P11388 | DNA topoisomerase 2-alpha, EC 5.6.2.2 | 1 | Nucleoplasm | Eukaryota | Leukoencephalopathy, Progressive Multifocal; Carcinoma, Merkel Cell; Hepatitis, Viral, Human |
| TUBA4A | P68366 | Tubulin alpha-4A chain | 4 | Cytoskeleton | Eukaryota | AIDS Dementia Complex; Leukoencephalopathy, Progressive Multifocal |
| TUBB | P07437 | Tubulin beta chain | —— | Cytoplasm, cytoskeleton | Eukaryota | —— |
| TYMS | P04818 | Thymidylate synthase, TS, TSase, EC 2.1.1.45 | 1 | Nucleus | Euk + Bac | Sarcoma, Kaposi; Influenza, Human; Hepatitis |
| Gene Symbol | Uniprot ID | Protein Name | Drug Information | Recognized Target | SCG Virus Disease(S) Median Score | Associated Virus Disease(s) Name (Only List the Top 3) * | Evolutionary Age | Pubmed Number a |
|---|---|---|---|---|---|---|---|---|
| ADRA2A | P08913 | Alpha-2A adrenergic receptor | known | Yes | 3 | AIDS Dementia Complex; Hepatitis | Eumetazoa | 13 |
| ADRB2 | P07550 | Beta-2 adrenergic receptor | known | Yes | 3 | Leukoencephalopathy, Progressive Multifocal; Paramyxoviridae Infections; Picornaviridae Infections | Eumetazoa | 52 |
| ADRB3 | P13945 | Beta-3 adrenergic receptor | known | Yes | 3 | Hepatitis; AIDS Dementia Complex; Picornaviridae Infections | Eumetazoa | 1 |
| AGER | Q15109 | Advanced glycosylation end product-specific receptor | unknown | No | 3 | Leukoencephalopathy, Progressive Multifocal; Hepatitis; AIDS Dementia Complex | Eumetazoa | 27 |
| AGTR1 | P30556 | Type-1 angiotensin II receptor | known | Yes | 3 | Leukoencephalopathy, Progressive Multifocal; Paramyxoviridae Infections; Hepatitis | Eumetazoa | 3 |
| ASIC1 | P78348 | Acid-sensing ion channel 1, ASIC1 | known | Yes | 3 | Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 3 |
| BIN1 | O00499 | Myc box-dependent-interacting protein 1 | unknown | No | 6 | Fatigue Syndrome, Chronic; AIDS Dementia Complex | Eumetazoa | 14 |
| BSG | P35613 | Basigin | unknown | No | 3 | Hepatitis; West Nile Fever; Epstein–Barr Virus Infections | Eumetazoa | 19 |
| CALHM1 | Q8IU99 | Calcium homeostasis modulator protein 1 | unknown | No | 9 | AIDS Dementia Complex | Eumetazoa | 1 |
| CD209 | Q9NNX6 | CD209 antigen | unknown | Yes | 3 | West Nile Fever; Hepatitis; Hemorrhagic Fever, Ebola | Eumetazoa | 65 |
| CD86 | P42081 | T-lymphocyte activation antigen CD86 | known | Yes | 3 | Respiratory Syncytial Virus Infections; Picornaviridae Infections; Hepatitis D | Eumetazoa | 758 |
| CRHR2 | Q13324 | Corticotropin-releasing factor receptor 2 | unknown | Yes | 3 | Hepatitis; Fatigue Syndrome, Chronic; West Nile Fever | Eumetazoa | 4 |
| DRD4 | P21917 | D(4) dopamine receptor | known | Yes | 3 | AIDS Dementia Complex | Eumetazoa | 5 |
| DRP2 | Q13474 | Dystrophin-related protein 2 | unknown | No | 3 | AIDS Dementia Complex | Eumetazoa | 1 |
| FAS | P25445 | Tumor necrosis factor receptor superfamily member 6 | unknown | Yes | 3 | Picornaviridae Infections; AIDS Dementia Complex; Carcinoma, Merkel Cell | Eumetazoa | 1588 |
| GHSR | Q92847 | Growth hormone secretagogue receptor type 1 | known | Yes | 3 | Carcinoma, Merkel Cell | Eumetazoa | 7 |
| GPC5 | P78333 | Glypican-5 | unknown | No | 3 | Leukoencephalopathy, Progressive Multifocal; | Eumetazoa | 1 |
| GPR65 | Q8IYL9 | Psychosine receptor | unknown | No | 6 | Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 1 |
| GRM4 | Q14833 | Metabotropic glutamate receptor 4 | known | Yes | 3 | Leukoencephalopathy, Progressive Multifocal; Hepatitis; Picornaviridae Infections | Eumetazoa | 1 |
| GRPR | P30550 | Gastrin-releasing peptide receptor | unknown | Yes | 3 | AIDS Dementia Complex | Eumetazoa | 1 |
| IL1R1 | P14778 | Interleukin-1 receptor type 1 | known | Yes | 3 | Leukoencephalopathy, Progressive Multifocal; Picornaviridae Infections; HIV Infections | Eumetazoa | 17 |
| IL1RL1 | Q01638 | Interleukin-1 receptor-like 1 | unknown | No | 3 | Influenza, Human; Hepatitis D; West Nile Fever | Eumetazoa | 31 |
| KCNQ1 | P51787 | Potassium voltage-gated channel subfamily KQT member 1 | known | Yes | 3 | Picornaviridae Infections; AIDS Dementia Complex; Measles | Eumetazoa | 5 |
| LGR5 | O75473 | Leucine-rich repeat-containing G-protein coupled receptor 5 | unknown | Yes | 3 | HIV Infections; Influenza, Human; Paramyxoviridae Infections | Eumetazoa | 25 |
| MAG | P20916 | Myelin-associated glycoprotein | unknown | No | 3 | Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 100 |
| NPSR1 | Q6W5P4 | Neuropeptide S receptor | known | Yes | 3 | Hepatitis; Respiratory Syncytial Virus Infections; Influenza, Human | Eumetazoa | 1 |
| PARD6A | Q9NPB6 | Partitioning defective 6 homolog alpha | unknown | No | 3 | AIDS Dementia Complex | Eumetazoa | 2 |
| PERP | Q96FX8 | p53 apoptosis effector related to PMP-22 | unknown | No | 3 | Carcinoma, Merkel Cell; Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 5 |
| RGS7 | P49802 | Regulator of G-protein signaling 7 | unknown | No | 3 | Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 1 |
| RHOU | Q7L0Q8 | Rho-related GTP-binding protein RhoU | unknown | No | 3 | Carcinoma, Merkel Cell | Eumetazoa | 4 |
| SGCA | Q16586 | Alpha-sarcoglycan, Alpha-SG | unknown | No | 14 | Fatigue Syndrome, Chronic | Eumetazoa | 7 |
| SHB | Q15464 | SH2 domain-containing adapter protein B | unknown | No | 3 | AIDS Dementia Complex | Eumetazoa | 19 |
| SYNE2 | Q8WXH0 | Nesprin-2 | unknown | No | 7 | Fatigue Syndrome, Chronic; | Eumetazoa | 4 |
| TGFBR1 | P36897 | TGF-beta receptor type-1 | known | Yes | 3 | Hepatitis; Hepatitis B; Carcinoma, Merkel Cell | Eumetazoa | 29 |
| TNFSF10 | P50591 | Tumor necrosis factor ligand superfamily member 10 | unknown | No | 3 | Hepatitis D; HIV Infections; Leukoencephalopathy, Progressive Multifocal | Eumetazoa | 224 |
| Gene Symbol | PDB ID | Protein Name | Resolution (Å) | Ligand(s) a | Number of Pockets | Number of Druggable Pockets (Score ≥ 0.5) | Best Druggable Pocket Score |
|---|---|---|---|---|---|---|---|
| AGER | 3O3U | Advanced glycosylation end product-specific receptor | 1.50 | MLR, SO4 | 24 | 13 | 1.00 |
| BIN1 | 2FIC | Myc box-dependent-interacting protein 1 | 1.99 | XE | 11 | 7 | 0.91 |
| BSG | 3I84 | Basigin | 2.00 | CL | 2 | 0 | —— |
| CD209 | 2XR6 | CD209 antigen | 1.35 | 07B, MAN, AE9, CA, CL | 2 | 2 | 0.78 |
| CRHR2 | 3N93-AB | Corticotropin-releasing factor receptor 2 | 2.50 | MAL, GOL | 41 | 18 | 1.00 |
| FAS | 3TJE-F | Tumor necrosis factor receptor superfamily member 6 | 1.93 | CD, EDO, CL | 4 | 1 | 0.84 |
| IL1RL1 | 4KC3-B | Interleukin-1 receptor-like 1 | 3.27 | NAG, MSE | 11 | 7 | 0.98 |
| LGR5 | 4UFR-AC | Leucine-rich repeat-containing G-protein coupled receptor 5 | 2.20 | NAG, CL | 34 | 21 | 0.99 |
| PARD6A | 1WMH-B | Partitioning defective 6 homolog alpha | 1.50 | —— | 4 | 3 | 0.99 |
| RGS7 | 2A72 | Regulator of G-protein signaling 7 | 2.00 | CL | 7 | 2 | 0.60 |
| RHOU | 2Q3H | Rho-related GTP-binding protein RhoU | 1.73 | GDP, MG | 6 | 1 | 0.93 |
| TNFSF10 | 1DG6-A | Tumor necrosis factor ligand superfamily member 10 | 1.30 | ZN, CL | 2 | 1 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhang, Q.-Y.; Chu, X.-Y.; Quan, Y.; Lv, B.-M.; Zhang, H.-Y. Facilitating Antiviral Drug Discovery Using Genetic and Evolutionary Knowledge. Viruses 2021, 13, 2117. https://doi.org/10.3390/v13112117
Xu X, Zhang Q-Y, Chu X-Y, Quan Y, Lv B-M, Zhang H-Y. Facilitating Antiviral Drug Discovery Using Genetic and Evolutionary Knowledge. Viruses. 2021; 13(11):2117. https://doi.org/10.3390/v13112117
Chicago/Turabian StyleXu, Xuan, Qing-Ye Zhang, Xin-Yi Chu, Yuan Quan, Bo-Min Lv, and Hong-Yu Zhang. 2021. "Facilitating Antiviral Drug Discovery Using Genetic and Evolutionary Knowledge" Viruses 13, no. 11: 2117. https://doi.org/10.3390/v13112117
APA StyleXu, X., Zhang, Q.-Y., Chu, X.-Y., Quan, Y., Lv, B.-M., & Zhang, H.-Y. (2021). Facilitating Antiviral Drug Discovery Using Genetic and Evolutionary Knowledge. Viruses, 13(11), 2117. https://doi.org/10.3390/v13112117







