Species-Specific Inhibition of Necroptosis by HCMV UL36
Abstract
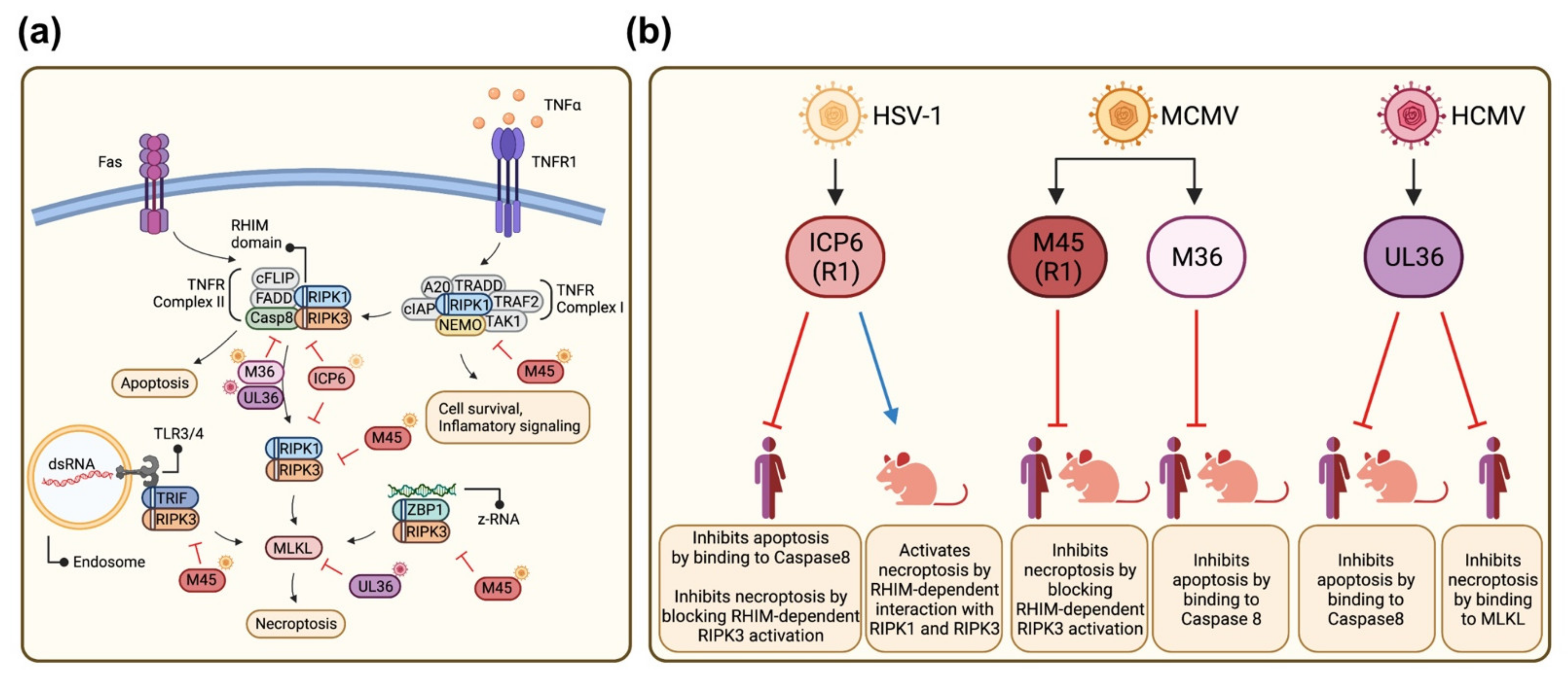
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Antibodies
2.3. Plasmids
2.4. Transfection
2.5. Cell Viability Assay
2.6. Immunodetection
3. Results
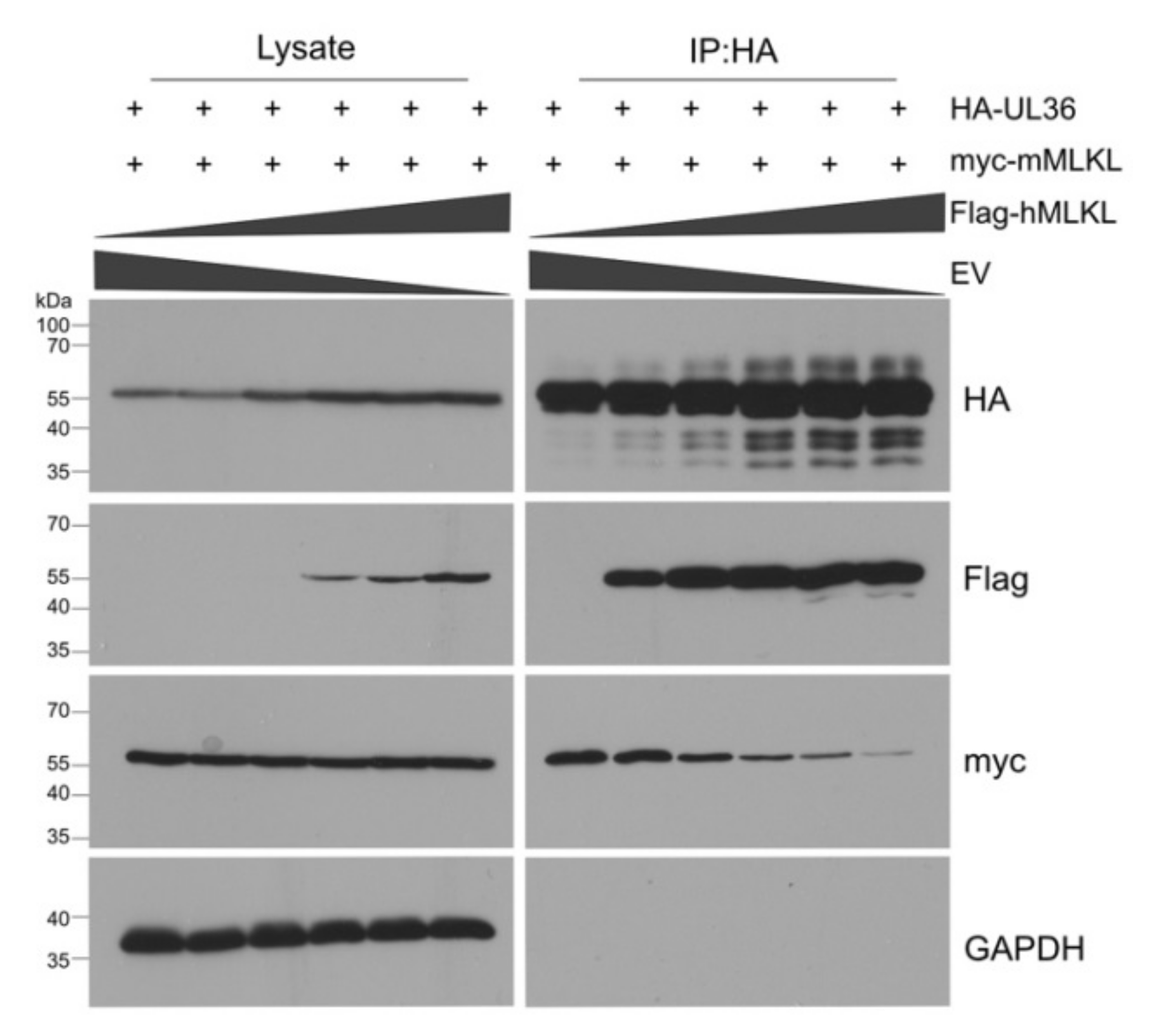
3.1. HCMV UL36 Interacts with Human and Murine MLKL
3.2. UL36 Interacts with the N-Terminal Domain of MLKLs
3.3. UL36 Has a Higher Affinity to hMLKL Than to mMLKL
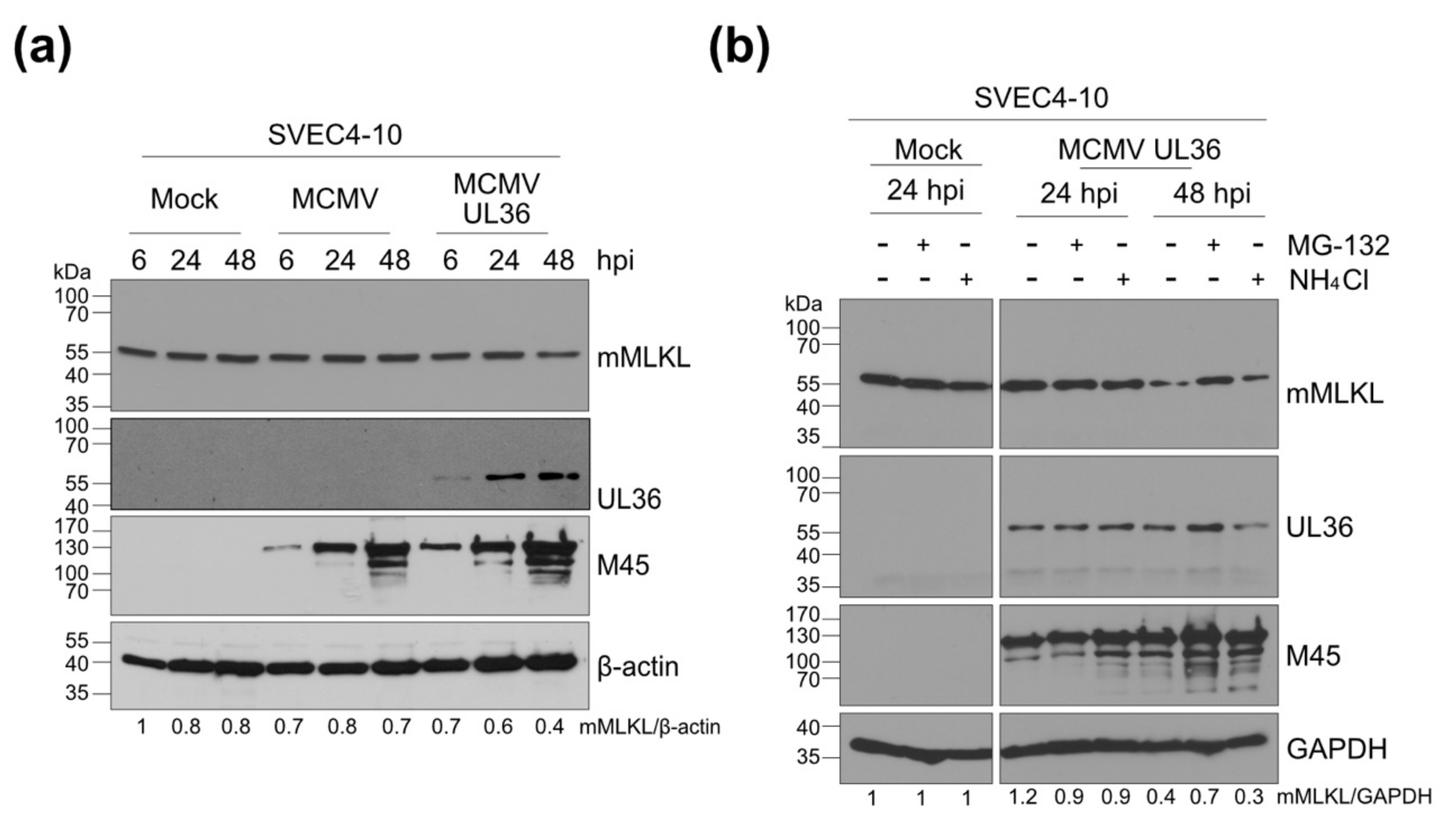
3.4. UL36 Promotes Proteasomal Degradation of mMLKL in Infected Cells
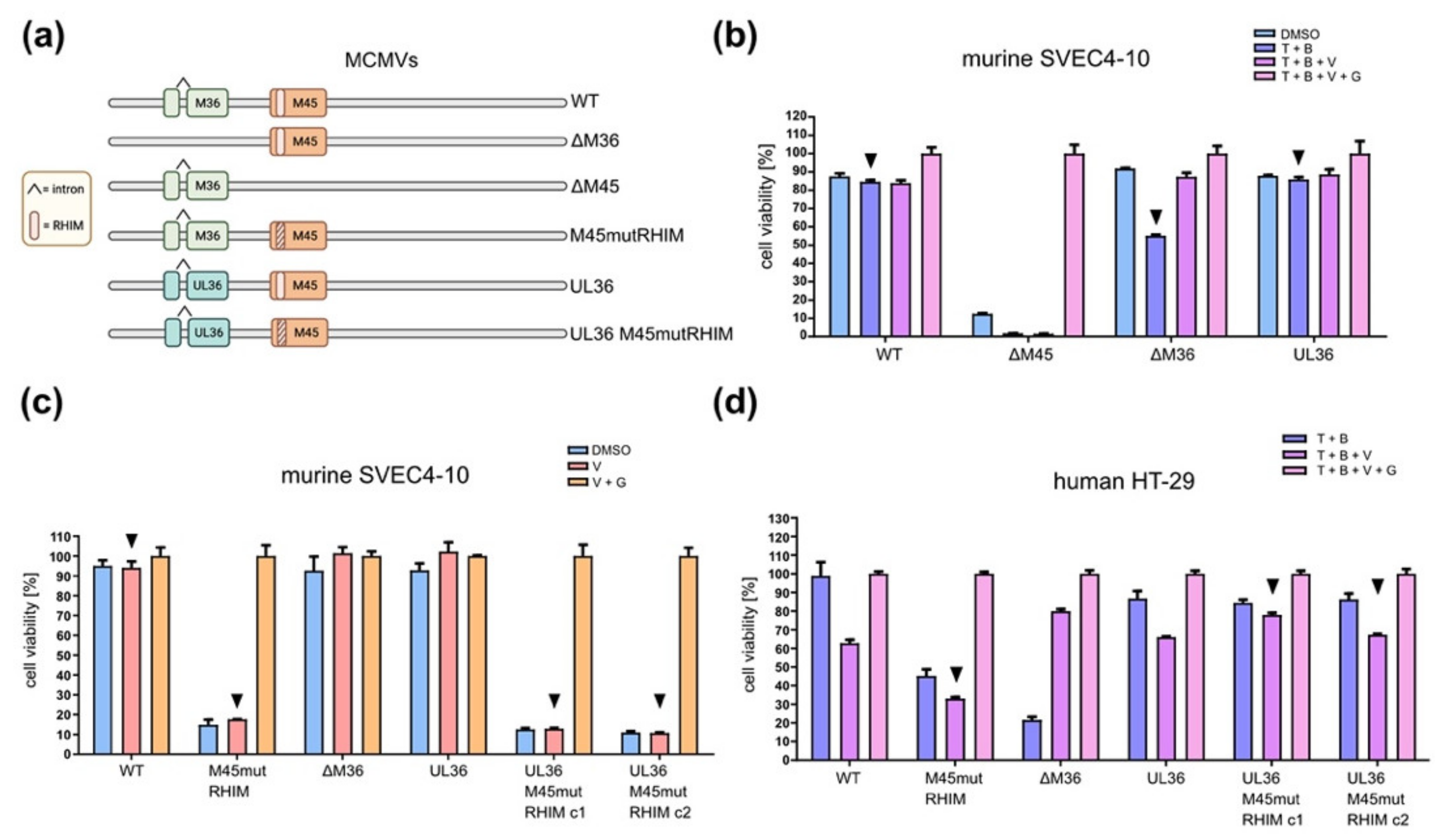
3.5. A Chimeric MCMV-UL36 Virus Prevents Necroptosis in Humans but Not in Murine Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pellet, P.E.; Roizman, B. Herpesviridiae. In Fields Virology, 6th ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1802–1822. [Google Scholar]
- Arvin, A.; Campadelli-Fiume, G.; Mocarski, E.; Moore, P.S.; Roizman, B.; Whitley, R.; Yamanishi, K. (Eds.) Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- O’Connor, C.M.; Sen, G.C. Innate Immune Responses to Herpesvirus Infection. Cells 2021, 10, 2122. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Viral Subversion of the Host Protein Synthesis Machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Luo, H. Interplay between the Virus and the Ubiquitin-Proteasome System: Molecular Mechanism of Viral Pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Imre, G. Cell Death Signalling in Virus Infection. Cell. Signal. 2020, 76, 109772. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular Mechanisms of Cell Death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Orzalli, M.H.; Kagan, J.C. Apoptosis and Necroptosis as Host Defense Strategies to Prevent Viral Infection. Trends Cell Biol. 2017, 27, 800–809. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, Pyroptosis and Apoptosis: An Intricate Game of Cell Death. Cell. Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M.F. Tumour Necrosis Factor Signalling in Health and Disease. F1000Research 2019, 8, F1000Faculty Rev-111. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-W.; Seo, J.; Jeong, M.; Lee, S.; Song, J. The Roles of FADD in Extrinsic Apoptosis and Necroptosis. BMB Rep. 2012, 45, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled Demolition at the Cellular Level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Orozco, S.; Oberst, A. RIPK3 in Cell Death and Inflammation: The Good, the Bad, and the Ugly. Immunol. Rev. 2017, 277, 102–112. [Google Scholar] [CrossRef]
- Grootjans, S.; Vanden Berghe, T.; Vandenabeele, P. Initiation and Execution Mechanisms of Necroptosis: An Overview. Cell Death Differ. 2017, 24, 1184–1195. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; He, W.; Sun, L. Necrosome Core Machinery: MLKL. Cell. Mol. Life Sci. CMLS 2016, 73, 2153–2163. [Google Scholar] [CrossRef]
- He, S.; Han, J. Manipulation of Host Cell Death Pathways by Herpes Simplex Virus. Curr. Top. Microbiol. Immunol. 2020. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Guo, H.; Kaiser, W.J. Necroptosis: The Trojan Horse in Cell Autonomous Antiviral Host Defense. Virology 2015, 479–480, 160–166. [Google Scholar] [CrossRef]
- Brune, W.; Andoniou, C.E. Die Another Day: Inhibition of Cell Death Pathways by Cytomegalovirus. Viruses 2017, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Omoto, S.; Harris, P.A.; Finger, J.N.; Bertin, J.; Gough, P.J.; Kaiser, W.J.; Mocarski, E.S. Herpes Simplex Virus Suppresses Necroptosis in Human Cells. Cell Host Microbe 2015, 17, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, S.-Q.; Liang, Y.; Zhou, X.; Chen, W.; Li, L.; Wu, J.; Zhuang, Q.; Chen, C.; Li, J.; et al. RIP1/RIP3 Binding to HSV-1 ICP6 Initiates Necroptosis to Restrict Virus Propagation in Mice. Cell Host Microbe 2015, 17, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Sasseville, A.M.-J.; Chabaud, S.; Massie, B.; Siegel, R.M.; Langelier, Y. The Ribonucleotide Reductase R1 Subunits of Herpes Simplex Virus Types 1 and 2 Protect Cells against TNFα- and FasL-Induced Apoptosis by Interacting with Caspase-8. Apoptosis Int. J. Program. Cell Death 2011, 16, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, Y.; Chen, Q.; Su, C.; Zhang, Z.; Yang, C.; Hu, Z.; Hou, J.; Zhou, J.; Gong, L.; et al. Herpes Simplex Virus 1 (HSV-1) and HSV-2 Mediate Species-Specific Modulations of Programmed Necrosis through the Viral Ribonucleotide Reductase Large Subunit R1. J. Virol. 2016, 90, 1088–1095. [Google Scholar] [CrossRef]
- Skaletskaya, A.; Bartle, L.M.; Chittenden, T.; McCormick, A.L.; Mocarski, E.S.; Goldmacher, V.S. A Cytomegalovirus-Encoded Inhibitor of Apoptosis That Suppresses Caspase-8 Activation. Proc. Natl. Acad. Sci. USA 2001, 98, 7829–7834. [Google Scholar] [CrossRef] [PubMed]
- Fletcher-Etherington, A.; Nobre, L.; Nightingale, K.; Antrobus, R.; Nichols, J.; Davison, A.J.; Stanton, R.J.; Weekes, M.P. Human Cytomegalovirus Protein PUL36: A Dual Cell Death Pathway Inhibitor. Proc. Natl. Acad. Sci. USA 2020, 117, 18771–18779. [Google Scholar] [CrossRef] [PubMed]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. DAI Complexes with RIP3 to Mediate Virus-Induced Programmed Necrosis That Is Targeted by Murine Cytomegalovirus vIRA. Cell Host Microbe 2012, 11, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. Cytomegalovirus M45 Cell Death Suppression Requires Receptor-Interacting Protein (RIP) Homotypic Interaction Motif (RHIM)-Dependent Interaction with RIP1. J. Biol. Chem. 2008, 283, 16966–16970. [Google Scholar] [CrossRef]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. Virus Inhibition of RIP3-Dependent Necrosis. Cell Host Microbe 2010, 7, 302–313. [Google Scholar] [CrossRef]
- Ménard, C.; Wagner, M.; Ruzsics, Z.; Holak, K.; Brune, W.; Campbell, A.E.; Koszinowski, U.H. Role of Murine Cytomegalovirus US22 Gene Family Members in Replication in Macrophages. J. Virol. 2003, 77, 5557–5570. [Google Scholar] [CrossRef] [PubMed]
- Omoto, S.; Guo, H.; Talekar, G.R.; Roback, L.; Kaiser, W.J.; Mocarski, E.S. Suppression of RIP3-Dependent Necroptosis by Human Cytomegalovirus. J. Biol. Chem. 2015, 290, 11635–11648. [Google Scholar] [CrossRef] [PubMed]
- Poncet, D.; Larochette, N.; Pauleau, A.-L.; Boya, P.; Jalil, A.-A.; Cartron, P.-F.; Vallette, F.; Schnebelen, C.; Bartle, L.M.; Skaletskaya, A.; et al. An Anti-Apoptotic Viral Protein That Recruits Bax to Mitochondria *. J. Biol. Chem. 2004, 279, 22605–22614. [Google Scholar] [CrossRef]
- Arnoult, D.; Bartle, L.M.; Skaletskaya, A.; Poncet, D.; Zamzami, N.; Park, P.U.; Sharpe, J.; Youle, R.J.; Goldmacher, V.S. Cytomegalovirus Cell Death Suppressor vMIA Blocks Bax- but Not Bak-Mediated Apoptosis by Binding and Sequestering Bax at Mitochondria. Proc. Natl. Acad. Sci. USA 2004, 101, 7988–7993. [Google Scholar] [CrossRef] [PubMed]
- Goldmacher, V.S.; Bartle, L.M.; Skaletskaya, A.; Dionne, C.A.; Kedersha, N.L.; Vater, C.A.; Han, J.; Lutz, R.J.; Watanabe, S.; McFarland, E.D.C.; et al. A Cytomegalovirus-Encoded Mitochondria-Localized Inhibitor of Apoptosis Structurally Unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 1999, 96, 12536–12541. [Google Scholar] [CrossRef] [PubMed]
- Karbowski, M.; Norris, K.L.; Cleland, M.M.; Jeong, S.-Y.; Youle, R.J. Role of Bax and Bak in Mitochondrial Morphogenesis. Nature 2006, 443, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Norris, K.L.; Youle, R.J. Cytomegalovirus Proteins vMIA and m38.5 Link Mitochondrial Morphogenesis to Bcl-2 Family Proteins. J. Virol. 2008, 82, 6232–6243. [Google Scholar] [CrossRef] [PubMed]
- Cam, M.; Handke, W.; Picard-Maureau, M.; Brune, W. Cytomegaloviruses Inhibit Bak- and Bax-Mediated Apoptosis with Two Separate Viral Proteins. Cell Death Differ. 2010, 17, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Schumacher, U.; Simic, H.; Voigt, S.; Brune, W. Murine Cytomegalovirus m38.5 Protein Inhibits Bax-Mediated Cell Death. J. Virol. 2008, 82, 4812–4822. [Google Scholar] [CrossRef] [PubMed]
- Schmiedeke, J.K.; Hartmann, A.-K.; Ruckenbrod, T.; Stassen, M.; Reddehase, M.J.; Lemmermann, N.A. The Anti-Apoptotic Murine Cytomegalovirus Protein vMIA-m38.5 Induces Mast Cell Degranulation. Front. Cell. Infect. Microbiol. 2020, 10, 439. [Google Scholar] [CrossRef] [PubMed]
- Handke, W.; Luig, C.; Popovic, B.; Krmpotic, A.; Jonjic, S.; Brune, W. Viral Inhibition of BAK Promotes Murine Cytomegalovirus Dissemination to Salivary Glands. J. Virol. 2013, 87, 3592–3596. [Google Scholar] [CrossRef] [PubMed]
- McCormick, A.L.; Skaletskaya, A.; Barry, P.A.; Mocarski, E.S.; Goldmacher, V.S. Differential Function and Expression of the Viral Inhibitor of Caspase 8-Induced Apoptosis (vICA) and the Viral Mitochondria-Localized Inhibitor of Apoptosis (vMIA) Cell Death Suppressors Conserved in Primate and Rodent Cytomegaloviruses. Virology 2003, 316, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.Z.; Kasmapour, B.; Plaza-Sirvent, C.; Bajagic, M.; Casalegno Garduño, R.; Borkner, L.; Lenac Roviš, T.; Scrima, A.; Jonjic, S.; Schmitz, I.; et al. UL36 Rescues Apoptosis Inhibition and In Vivo Replication of a Chimeric MCMV Lacking the M36 Gene. Front. Cell. Infect. Microbiol. 2017, 7, 312. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.M.; Levine, A.J. P53 Alteration Is a Common Event in the Spontaneous Immortalization of Primary BALB/c Murine Embryo Fibroblasts. Genes Dev. 1991, 5, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.; Krause, J.; Prager, A.; Mitrovic, M.; Jonjic, S.; Koszinowski, U.H.; Adler, B. Virus Progeny of Murine Cytomegalovirus Bacterial Artificial Chromosome PSM3fr Show Reduced Growth in Salivary Glands Due to a Fixed Mutation of MCK-2. J. Virol. 2011, 85, 10346–10353. [Google Scholar] [CrossRef] [PubMed]
- Krause, E.; de Graaf, M.; Fliss, P.M.; Dölken, L.; Brune, W. Murine Cytomegalovirus Virion-Associated Protein M45 Mediates Rapid NF-κB Activation after Infection. J. Virol. 2014, 88, 9963–9975. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En Passant Mutagenesis: A Two Step Markerless Red Recombination System. Methods Mol. Biol. Clifton NJ 2010, 634, 421–430. [Google Scholar] [CrossRef]
- Brune, W.; Hengel, H.; Koszinowski, U.H. A Mouse Model for Cytomegalovirus Infection. 2001. Available online: https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/0471142735.im1907s43 (accessed on 19 October 2021). [CrossRef] [PubMed]
- Kangro, H.O.; Mahy, B.W.J. Virology Methods Manual; Academic Press: San Diego, CA, USA, 1996. [Google Scholar] [CrossRef]
- Patterson, C.E.; Shenk, T. Human Cytomegalovirus UL36 Protein Is Dispensable for Viral Replication in Cultured Cells. J. Virol. 1999, 73, 7126–7131. [Google Scholar] [CrossRef] [PubMed]
- Hüttmann, J.; Krause, E.; Schommartz, T.; Brune, W. Functional Comparison of Molluscum Contagiosum Virus VFLIP MC159 with Murine Cytomegalovirus M36/vICA and M45/vIRA Proteins. J. Virol. 2015, 90, 2895–2905. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Petrie, E.J.; Sandow, J.J.; Jacobsen, A.V.; Smith, B.J.; Griffin, M.D.W.; Lucet, I.S.; Dai, W.; Young, S.N.; Tanzer, M.C.; Wardak, A.; et al. Conformational Switching of the Pseudokinase Domain Promotes Human MLKL Tetramerization and Cell Death by Necroptosis. Nat. Commun. 2018, 9, 2422. [Google Scholar] [CrossRef]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.-C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.-G.; Liu, Z.-G. Plasma Membrane Translocation of Trimerized MLKL Protein Is Required for TNF-Induced Necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, J.M.; Tanzer, M.C.; Lucet, I.S.; Young, S.N.; Spall, S.K.; Sharma, P.; Pierotti, C.; Garnier, J.-M.; Dobson, R.C.J.; Webb, A.I.; et al. Activation of the Pseudokinase MLKL Unleashes the Four-Helix Bundle Domain to Induce Membrane Localization and Necroptotic Cell Death. Proc. Natl. Acad. Sci. USA 2014, 111, 15072–15077. [Google Scholar] [CrossRef]
- Petrie, E.J.; Birkinshaw, R.W.; Koide, A.; Denbaum, E.; Hildebrand, J.M.; Garnish, S.E.; Davies, K.A.; Sandow, J.J.; Samson, A.L.; Gavin, X.; et al. Identification of MLKL Membrane Translocation as a Checkpoint in Necroptotic Cell Death Using Monobodies. Proc. Natl. Acad. Sci. USA 2020, 117, 8468–8475. [Google Scholar] [CrossRef]
- Rodriguez, D.A.; Weinlich, R.; Brown, S.; Guy, C.; Fitzgerald, P.; Dillon, C.P.; Oberst, A.; Quarato, G.; Low, J.; Cripps, J.G.; et al. Characterization of RIPK3-Mediated Phosphorylation of the Activation Loop of MLKL during Necroptosis. Cell Death Differ. 2016, 23, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Brune, W.; Ménard, C.; Heesemann, J.; Koszinowski, U.H. A Ribonucleotide Reductase Homolog of Cytomegalovirus and Endothelial Cell Tropism. Science 2001, 291, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Daley-Bauer, L.P.; Roback, L.; Crosby, L.N.; McCormick, A.L.; Feng, Y.; Kaiser, W.J.; Mocarski, E.S. Mouse Cytomegalovirus M36 and M45 Death Suppressors Cooperate to Prevent Inflammation Resulting from Antiviral Programmed Cell Death Pathways. Proc. Natl. Acad. Sci. USA 2017, 114, E2786–E2795. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Brune, W. Induction of Apoptosis Limits Cytomegalovirus Cross-Species Infection. EMBO J. 2006, 25, 2634–2642. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, U.; Handke, W.; Jurak, I.; Brune, W. Mutations in the M112/M113-Coding Region Facilitate Murine Cytomegalovirus Replication in Human Cells. J. Virol. 2010, 84, 7994–8006. [Google Scholar] [CrossRef][Green Version]
- Ostermann, E.; Loroch, S.; Qian, Z.; Sickmann, A.; Wiebusch, L.; Brune, W. Activation of E2F-Dependent Transcription by the Mouse Cytomegalovirus M117 Protein Affects the Viral Host Range. PLoS Pathog. 2018, 14, e1007481. [Google Scholar] [CrossRef]
- Guo, H.; Kaiser, W.J.; Mocarski, E.S. Manipulation of Apoptosis and Necroptosis Signaling by Herpesviruses. Med. Microbiol. Immunol. 2015, 204, 439–448. [Google Scholar] [CrossRef]
- Mack, C.; Sickmann, A.; Lembo, D.; Brune, W. Inhibition of Proinflammatory and Innate Immune Signaling Pathways by a Cytomegalovirus RIP1-Interacting Protein. Proc. Natl. Acad. Sci. USA 2008, 105, 3094–3099. [Google Scholar] [CrossRef]
- Lembo, D.; Brune, W. Tinkering with a Viral Ribonucleotide Reductase. Trends Biochem. Sci. 2009, 34, 25–32. [Google Scholar] [CrossRef]
- Hahn, G.; Khan, H.; Baldanti, F.; Koszinowski, U.H.; Revello, M.G.; Gerna, G. The Human Cytomegalovirus Ribonucleotide Reductase Homolog UL45 Is Dispensable for Growth in Endothelial Cells, as Determined by a BAC-Cloned Clinical Isolate of Human Cytomegalovirus with Preserved Wild-Type Characteristics. J. Virol. 2002, 76, 9551–9555. [Google Scholar] [CrossRef]
- Muscolino, E.; Schmitz, R.; Loroch, S.; Caragliano, E.; Schneider, C.; Rizzato, M.; Kim, Y.-H.; Krause, E.; Juranić Lisnić, V.; Sickmann, A.; et al. Herpesviruses Induce Aggregation and Selective Autophagy of Host Signalling Proteins NEMO and RIPK1 as an Immune-Evasion Mechanism. Nat. Microbiol. 2020, 5, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Muscolino, E.; Luoto, L.-M.; Brune, W. Viral Induced Protein Aggregation: A Mechanism of Immune Evasion. Int. J. Mol. Sci. 2021, 22, 9624. [Google Scholar] [CrossRef] [PubMed]
- Fliss, P.M.; Jowers, T.P.; Brinkmann, M.M.; Holstermann, B.; Mack, C.; Dickinson, P.; Hohenberg, H.; Ghazal, P.; Brune, W. Viral Mediated Redirection of NEMO/IKKγ to Autophagosomes Curtails the Inflammatory Cascade. PLoS Pathog. 2012, 8, e1002517. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.M.; Oh, S.E.; Kim, Y.E.; Han, T.-H.; Ahn, J.-H. Cooperative Inhibition of RIP1-Mediated NF-κB Signaling by Cytomegalovirus-Encoded Deubiquitinase and Inactive Homolog of Cellular Ribonucleotide Reductase Large Subunit. PLoS Pathog. 2017, 13, e1006423. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, Z.; Li, L.; Zhong, C.-Q.; Zheng, X.; Wu, X.; Zhang, Y.; Ma, H.; Huang, D.; Li, W.; et al. Diverse Sequence Determinants Control Human and Mouse Receptor Interacting Protein 3 (RIP3) and Mixed Lineage Kinase Domain-Like (MLKL) Interaction in Necroptotic Signaling *. J. Biol. Chem. 2013, 288, 16247–16261. [Google Scholar] [CrossRef] [PubMed]
- Brune, W. Molecular Basis of Cytomegalovirus Host Species Specificity. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; Volume 1, pp. 322–329. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muscolino, E.; Castiglioni, C.; Brixel, R.; Frascaroli, G.; Brune, W. Species-Specific Inhibition of Necroptosis by HCMV UL36. Viruses 2021, 13, 2134. https://doi.org/10.3390/v13112134
Muscolino E, Castiglioni C, Brixel R, Frascaroli G, Brune W. Species-Specific Inhibition of Necroptosis by HCMV UL36. Viruses. 2021; 13(11):2134. https://doi.org/10.3390/v13112134
Chicago/Turabian StyleMuscolino, Elena, Claudia Castiglioni, Renke Brixel, Giada Frascaroli, and Wolfram Brune. 2021. "Species-Specific Inhibition of Necroptosis by HCMV UL36" Viruses 13, no. 11: 2134. https://doi.org/10.3390/v13112134
APA StyleMuscolino, E., Castiglioni, C., Brixel, R., Frascaroli, G., & Brune, W. (2021). Species-Specific Inhibition of Necroptosis by HCMV UL36. Viruses, 13(11), 2134. https://doi.org/10.3390/v13112134






