Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Virus, and Reagents
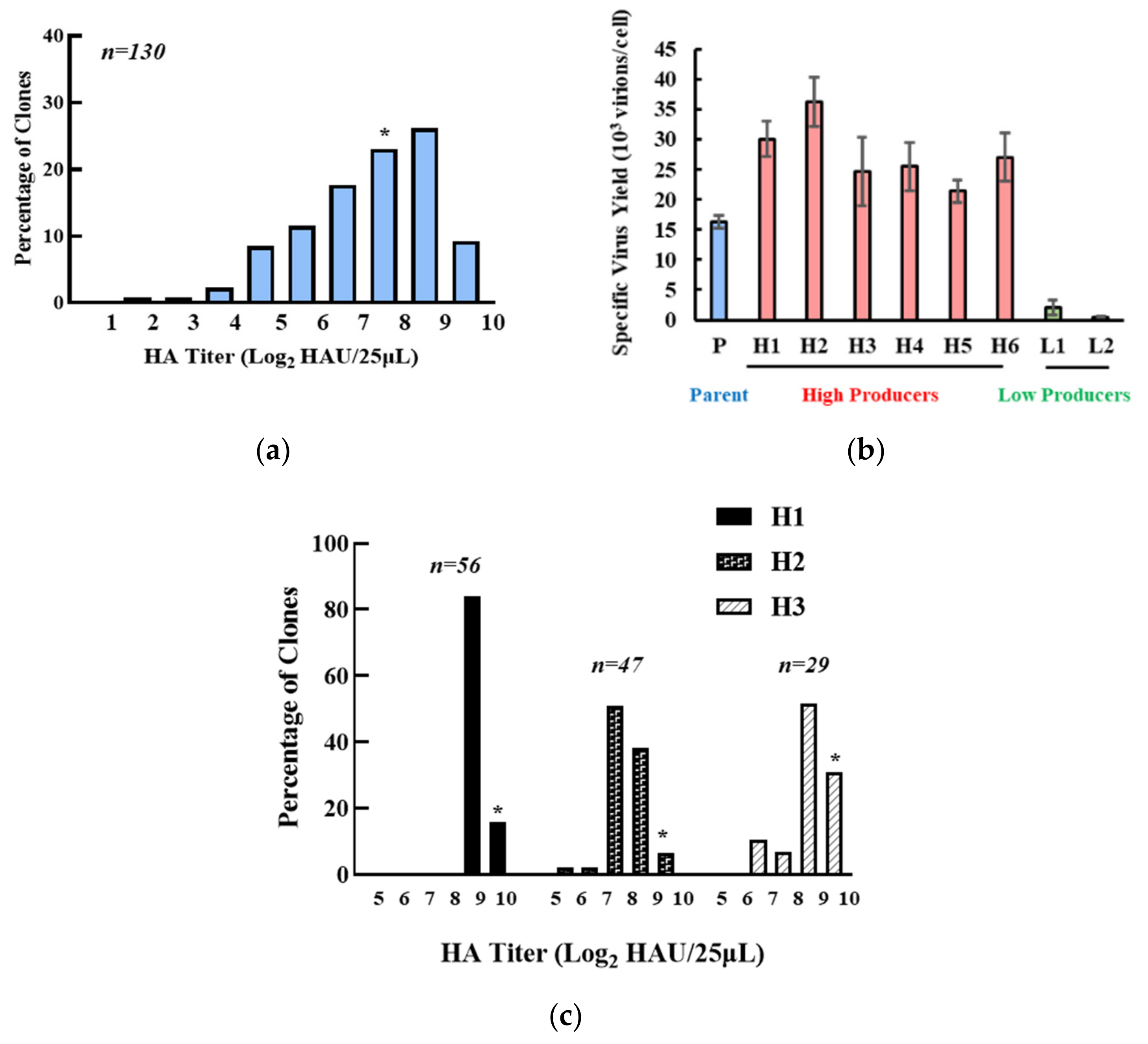
2.2. Single Cell Cloning
2.3. Virus Infection
2.4. Virus Quantification
2.5. Digoxigenin Sialic Acid (SA) Detection Assay
2.6. Analysis of Apoptosis during IAV Infection
2.7. RNA Sequencing by Illumina HiSeq and Transcriptome Analysis
2.8. Mitochondrial Protein Separation, Quantification, and Mitochondrial Membrane Potential (MMP) Assay
2.9. Statistical Analysis
3. Results
3.1. Isolation of the High IAV-Producing Clone
3.2. H1 Sustained Virus Production Longer in Synchronized Single-Cycle Infection
3.3. H1 Had Higher Viral Transcript Levels
3.4. Parallel Functional Responses of H1 and P to IAV Infection
3.5. Identifying Differentially Expressed Functional Classes between H1 and P
3.6. Differential Functional Enrichment in H1 Versus P Comparisons
3.6.1. Elevated Mitochondrial Activities in H1
3.6.2. Subdued Inflammatory and Antiviral Response in H1
4. Discussion
4.1. H1 and P Respond to Infection Similarly but Differ in Their Magnitude
4.2. H1 and P Both Exhibit Host Shutoff but H1 Retained Higher Activities
4.3. H1 Had Subdued Inflammatory and Antiviral Responses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Li, Y.T.; Linster, M.; Mendenhall, I.H.; Su, Y.C.F.; Smith, G.J.D. Avian influenza viruses in humans: Lessons from past outbreaks. Brit. Med. Bull. 2019, 132, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Barr, I.G.; Donis, R.O.; Katz, J.M.; McCauley, J.W.; Odagiri, T.; Trusheim, H.; Tsai, T.F.; Wentworth, D.E. Cell culture-derived influenza vaccines in the severe 2017–2018 epidemic season: A step towards improved influenza vaccine effectiveness. NPJ Vaccines 2018, 3, 44. [Google Scholar]
- Chu, C.; Lugovtsev, V.; Golding, H.; Betenbaugh, M.; Shiloach, J. Conversion of MDCK cell line to suspension culture by transfecting with human siat7e gene and its application for influenza virus production. Proc. Natl. Acad. Sci. USA 2009, 106, 14802–14807. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Peng, W.J.; Ye, Q.; Liu, X.P.; Zhao, L.; Fan, L.; Xia-Hou, K.; Jia, H.J.; Luo, J.; Zhou, L.T.; et al. Serum-Free Suspension Culture of MDCK Cells for Production of Influenza H1N1 Vaccines. PLoS ONE 2015, 10, e0141686. [Google Scholar] [CrossRef] [PubMed]
- Bissinger, T.; Fritsch, J.; Mihut, A.; Wu, Y.; Liu, X.; Genzel, Y.; Tan, W.S.; Reichl, U. Semi-perfusion cultures of suspension MDCK cells enable high cell concentrations and efficient influenza A virus production. Vaccine 2019, 37, 7003–7010. [Google Scholar] [CrossRef] [PubMed]
- Coronel, J.; Granicher, G.; Sandig, V.; Noll, T.; Genzel, Y.; Reichl, U. Application of an Inclined Settler for Cell Culture-Based Influenza A Virus Production in Perfusion Mode. Front. Bioeng. Biotechnol. 2020, 8, 672. [Google Scholar] [CrossRef]
- Vazquez-Ramirez, D.; Jordan, I.; Sandig, V.; Genzel, Y.; Reichl, U. High titer MVA and influenza A virus production using a hybrid fed-batch/perfusion strategy with an ATF system. Appl. Microbiol. Biotechnol. 2019, 103, 3025–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emma, P.; Kamen, A. Real-time monitoring of influenza virus production kinetics in HEK293 cell cultures. Biotechnol. Prog. 2013, 29, 275–284. [Google Scholar] [CrossRef]
- Rudiger, D.; Kupke, S.Y.; Laske, T.; Zmora, P.; Reichl, U. Multiscale modeling of influenza A virus replication in cell cultures predicts infection dynamics for highly different infection conditions. PLoS Comput. Biol. 2019, 15, e1006819. [Google Scholar] [CrossRef] [Green Version]
- Genzel, Y. Designing cell lines for viral vaccine production: Where do we stand? Biotechnol. J. 2015, 10, 728–740. [Google Scholar] [CrossRef]
- Park, S.J.; Kumar, M.; Kwon, H.I.; Seong, R.K.; Han, K.; Song, J.M.; Kim, C.J.; Choi, Y.K.; Shin, O.S. Dynamic changes in host gene expression associated with H5N8 avian influenza virus infection in mice. Sci. Rep. 2015, 5, 16512. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, K.; Liu, L.; Li, W.; Zhu, B.; Zhang, S.; Xu, P.; Liu, W.; Li, J. Global transcriptome analysis of H5N1 influenza virus-infected human cells. Hereditas 2019, 156, 10. [Google Scholar] [CrossRef] [Green Version]
- Dissanayake, T.K.; Schauble, S.; Mirhakkak, M.; Wu, W.L.; Ng, A.C.; Yip, C.C.Y.; Lopez, A.G.; Yeung, M.L.; Chan, K.H.; Yuen, K.Y.; et al. Comparative Transcriptomic Analysis of Rhinovirus and Influenza Virus Infection. Front. Microbiol. 2020, 11, 1580. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Zeng, H.; Kelly, S.M.; Tumpey, T.M.; Katze, M.G. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: Specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. mBio 2014, 5, e01102-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, P.; Ning, Z.; Hao, X.; Lin, X.; Zheng, Q.; Li, S. Comparative Analysis of Whole-Transcriptome RNA Expression in MDCK Cells Infected with the H3N2 and H5N1 Canine Influenza Viruses. Front. Cell Infect. Microbiol. 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalbfuss, B.; Knochlein, A.; Krober, T.; Reichl, U. Monitoring influenza virus content in vaccine production: Precise assays for the quantitation of hemagglutination and neuraminidase activity. Biologicals 2008, 36, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.H.; Schlesinger, R.W. Morphological and Quantitative Comparison between Infectious and Non-Infectious Forms of Influenza Virus. J. Exp. Med. 1954, 100, 203. [Google Scholar] [CrossRef] [Green Version]
- Karakus, U.; Crameri, M.; Lanz, C.; Yanguez, E. Propagation and Titration of Influenza Viruses. Methods Mol. Biol. 2018, 1836, 59–88. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.P.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J.; et al. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2017, 45, D466–D474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis, 2nd edition. Meas-Interdiscip. Res. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Lugovtsev, V.Y.; Melnyk, D.; Weir, J.P. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS ONE 2013, 8, e75014. [Google Scholar] [CrossRef]
- Nobusawa, E.; Aoyama, T.; Kato, H.; Suzuki, Y.; Tateno, Y.; Nakajima, K. Comparison of Complete Amino-Acid-Sequences and Receptor-Binding Properties among 13 Serotypes of Hemagglutinins of Influenza a-Viruses. Virology 1991, 182, 475–485. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C. Receptor Determinants of Human and Animal Influenza-Virus Isolates—Differences in Receptor Specificity of the Hemagglutinin-H-3 Based on Species of Origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Frensing, T.; Kupke, S.Y.; Bachmann, M.; Fritzsche, S.; Gallo-Ramirez, L.E.; Reichl, U. Influenza virus intracellular replication dynamics, release kinetics, and particle morphology during propagation in MDCK cells. Appl. Microbiol. Biotechnol. 2016, 100, 7181–7192. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Liu, S.; Goraya, M.U.; Maarouf, M.; Huang, S.; Chen, J.L. Host Immune Response to Influenza A Virus Infection. Front. Immunol. 2018, 9, 320. [Google Scholar] [CrossRef] [Green Version]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.M.; Kim, J.; Tenson, T.; Min, J.Y.; Kainov, D.E. Influenza Virus Infection, Interferon Response, Viral Counter-Response, and Apoptosis. Viruses 2017, 9, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bercovich-Kinori, A.; Tai, J.; Gelbart, I.A.; Shitrit, A.; Ben-Moshe, S.; Drori, Y.; Itzkovitz, S.; Mandelboim, M.; Stern-Ginossar, N. A systematic view on influenza induced host shutoff. eLife 2016, 5, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Zhao, W.M.; Lu, J.; Li, S.; Zhou, K.; Jiang, W.; Duan, X.F.; Fu, L.F.; Yu, B.L.; Cai, K.Q.; et al. Influenza virus matrix protein M1 interacts with SLD5 to block host cell cycle. Cell. Microbiol. 2019, 21, e13038. [Google Scholar] [CrossRef] [PubMed]
- Bogdanow, B.; Wang, X.; Eichelbaum, K.; Sadewasser, A.; Husic, I.; Paki, K.; Budt, M.; Hergeselle, M.; Vetter, B.; Hou, J.; et al. The dynamic proteome of influenza A virus infection identifies M segment splicing as a host range determinant. Nat. Commun. 2019, 10, 5518. [Google Scholar] [CrossRef] [Green Version]
- Fabozzi, G.; Oler, A.J.; Liu, P.C.; Chen, Y.; Mindaye, S.; Dolan, M.A.; Kenney, H.; Gucek, M.; Zhu, J.; Rabin, R.L.; et al. Strand-Specific Dual RNA Sequencing of Bronchial Epithelial Cells Infected with Influenza A/H3N2 Viruses Reveals Splicing of Gene Segment 6 and Novel Host-Virus Interactions. J. Virol. 2018, 92, e00518-18. [Google Scholar] [CrossRef] [Green Version]
- Phan, T.; Fay, E.J.; Lee, Z.; Aron, S.; Hu, W.S.; Langlois, R.A. Segment-specific kinetics of mRNA, cRNA and vRNA accumulation during influenza infection. J. Virol. 2021, 95, e02102-20. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, Y.; Lin, X.; Ren, C.; Zhao, J.; Wang, F.; Gao, X.; Xiao, R.; Zhao, L.; Chen, H.; et al. Influenza M2 protein regulates MAVS-mediated signaling pathway through interacting with MAVS and increasing ROS production. Autophagy 2019, 15, 1163–1181. [Google Scholar] [CrossRef]
- Levene, R.E.; Gaglia, M.M. Host Shutoff in Influenza A Virus: Many Means to an End. Viruses 2018, 10, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, M.T.; Horner, S.M. Flipping the script: Viral capitalization of RNA modifications. Brief. Funct. Genom. 2021, 20, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, N.M.; Van Eden, M.E.; Younan, P.; Lloyd, R.E. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: A novel mechanism for host translation shutoff. Mol. Cell Biol. 2004, 24, 1779–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgui, I.; Yanguez, E.; Sonenberg, N.; Nieto, A. Influenza virus mRNA translation revisited: Is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007, 81, 12427–12438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, D.T.; Serra-Moreno, R.; Singh, R.K.; Guatelli, J.C. BST-2/tetherin: A new component of the innate immune response to enveloped viruses. Trends Microbiol. 2010, 18, 388–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, E.; Oh, J.; Giao, N.Q.; Oh, S.; Park, S.H. Enhanced production of enveloped viruses in BST-2-deficient cell lines. Biotechnol. Bioeng. 2017, 114, 2289–2297. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yin, L.; Mei, S.; Li, J.; Xu, F.; Sun, H.; Liu, X.; Cen, S.; Liang, C.; Li, A.; et al. BST-2 restricts IAV release and is countered by the viral M2 protein. Biochem. J. 2017, 474, 717–730. [Google Scholar] [CrossRef]
- Hornung, V.; Hartmann, R.; Ablasser, A.; Hopfner, K.P. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014, 14, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Banerjee, S.; Wang, Y.; Goldstein, S.A.; Dong, B.; Gaughan, C.; Silverman, R.H.; Weiss, S.R. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. USA 2016, 113, 2241–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurokawa, C.; Iankov, I.D.; Galanis, E. A key anti-viral protein, RSAD2/VIPERIN, restricts the release of measles virus from infected cells. Virus Res. 2019, 263, 145–150. [Google Scholar] [CrossRef]
- Panayiotou, C.; Lindqvist, R.; Kurhade, C.; Vonderstein, K.; Pasto, J.; Edlund, K.; Upadhyay, A.S.; Overby, A.K. Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J. Virol. 2018, 92, e02054-17. [Google Scholar] [CrossRef] [Green Version]
- Hamamoto, I.; Takaku, H.; Tashiro, M.; Yamamoto, N. High yield production of influenza virus in Madin Darby canine kidney (MDCK) cells with stable knockdown of IRF7. PLoS ONE 2013, 8, e59892. [Google Scholar] [CrossRef]
- Kim, T.H.; Kern, C.; Zhou, H. Knockout of IRF7 Highlights its Modulator Function of Host Response Against Avian Influenza Virus and the Involvement of MAPK and TOR Signaling Pathways in Chicken. Genes 2020, 11, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Xin, Z.T.; Liang, Y.; Ly, H.; Liang, Y. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 2008, 82, 9880–9889. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.X.; Qin, Y.; Wu, H.; Chen, Y.; Wu, S.; Si, X.N.; Wang, H.; Wang, T.Y.; Zhong, X.Y.; Zhai, X.; et al. Pyrrolidine dithiocarbamate inhibits enterovirus 71 replication by down-regulating ubiquitin-proteasome system. Virus Res. 2015, 195, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Wiesener, N.; Zimmer, C.; Jarasch-Althof, N.; Wutzler, P.; Henke, A. Therapy of experimental influenza virus infection with pyrrolidine dithiocarbamate. Med. Microbiol. Immun. 2011, 200, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.G.; Xu, Y.; Zhang, Q.; Yang, F.; Yin, Z.; Wang, L.X.; Li, Q.F. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet. Microbiol. 2019, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Q.; Phan, T.; Hu, W.-S.; Liu, X.; Fan, L.; Tan, W.-S.; Zhao, L. Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses 2021, 13, 2200. https://doi.org/10.3390/v13112200
Ye Q, Phan T, Hu W-S, Liu X, Fan L, Tan W-S, Zhao L. Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses. 2021; 13(11):2200. https://doi.org/10.3390/v13112200
Chicago/Turabian StyleYe, Qian, Thu Phan, Wei-Shou Hu, Xuping Liu, Li Fan, Wen-Song Tan, and Liang Zhao. 2021. "Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells" Viruses 13, no. 11: 2200. https://doi.org/10.3390/v13112200
APA StyleYe, Q., Phan, T., Hu, W.-S., Liu, X., Fan, L., Tan, W.-S., & Zhao, L. (2021). Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses, 13(11), 2200. https://doi.org/10.3390/v13112200





