Constraint of Base Pairing on HDV Genome Evolution
Abstract
:1. Introduction
2. Materials and Methods
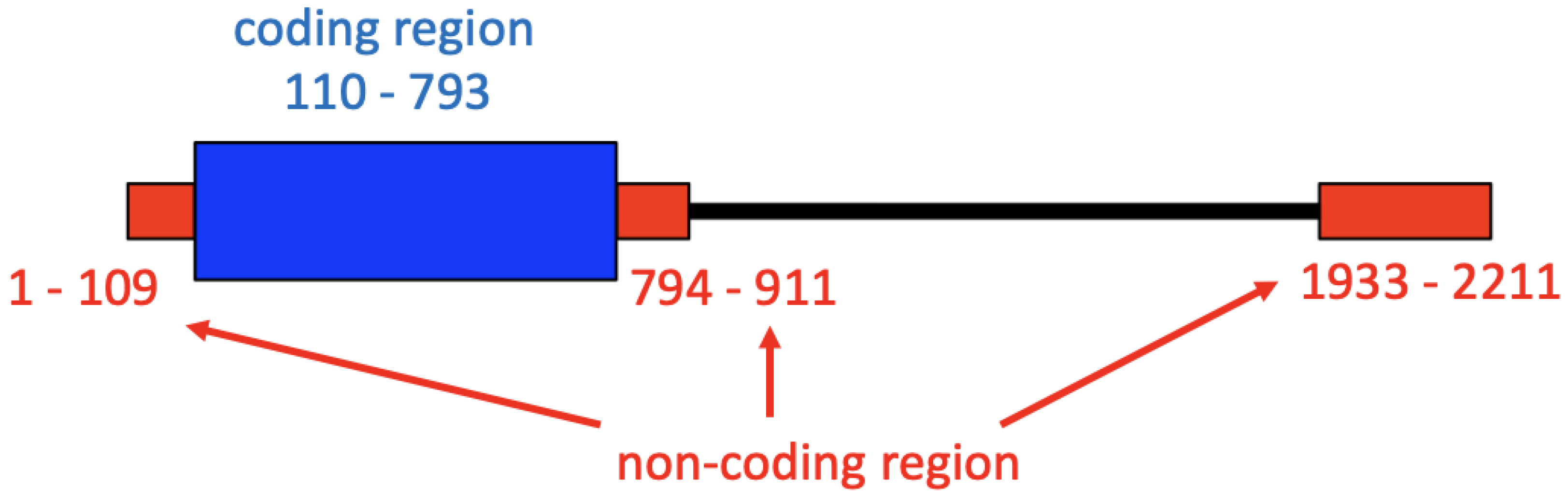
2.1. Alignment of HDV Genomes and L-HDAg Genes
2.2. Threshold for Base-Pairing Probability
2.3. Calculation of Evolutionary Distance
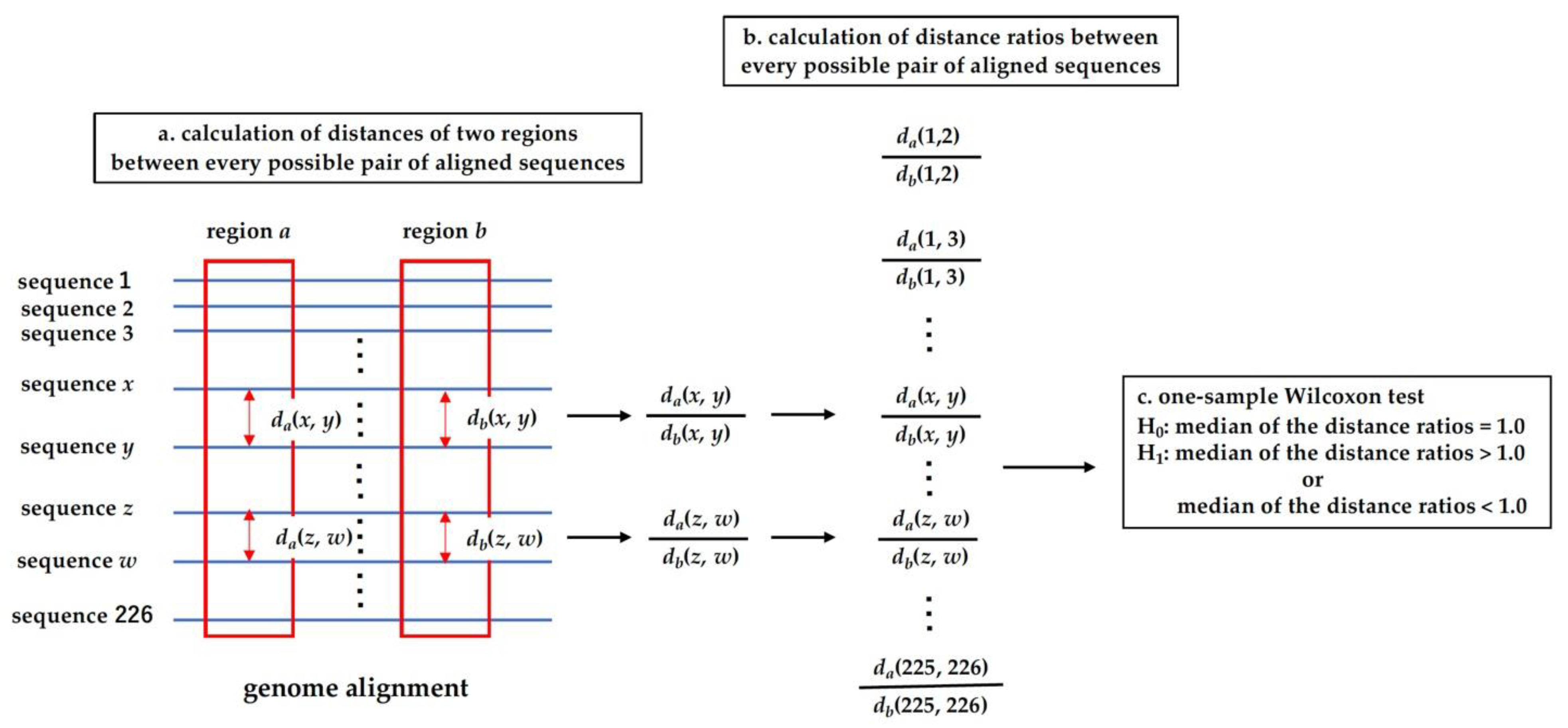
2.4. Analysis of Distance Ratio
3. Results
3.1. Comparison within Each Genotype
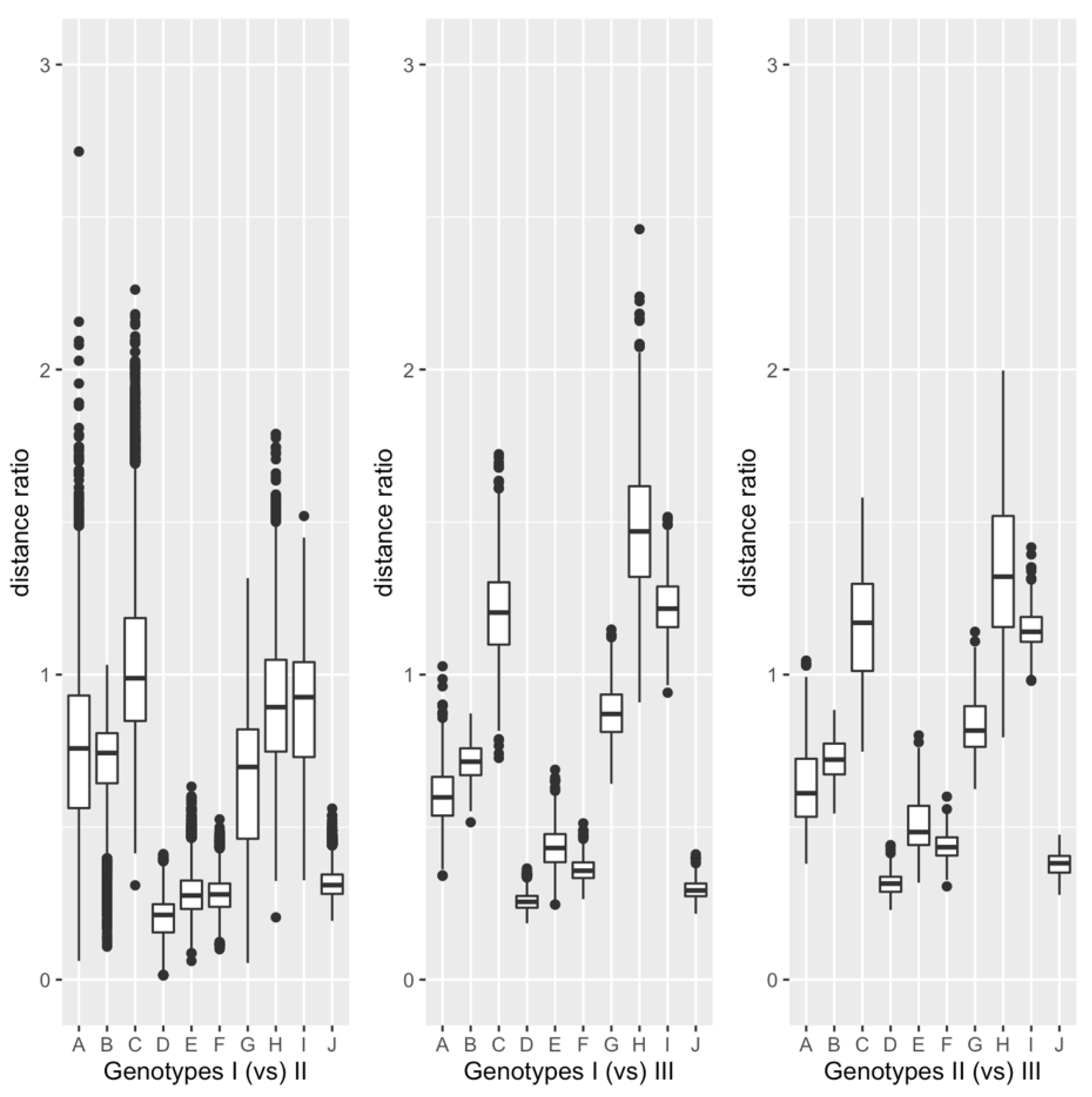
3.2. Comparison between Genotypes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Husa, P.; Linhartová, A.; Nemecek, V.; Husová, L. Hepatitis D. Acta Virol. 2005, 49, 219–225. [Google Scholar]
- Sureau, C.; Guerra, B.; Lanford, R.E. Role of the large hepatitis B virus envelope protein. J. Virol. 1993, 67, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Sureau, C. The role of the HBV envelope proteins in the HDV replication cycle. Curr. Top. Microbiol. Immunol. 2006, 307, 113–131. [Google Scholar] [CrossRef]
- Niro, G.A.; Smedile, A.; Andriulli, A.; Rizzetto, M.; Gerin, J.L.; Casey, J.L. The predominance of hepatitis delta virus genotype I among chronically infected Italian patients. Hepatology 1997, 25, 728–734. [Google Scholar] [CrossRef]
- Shakil, A.O.; Hadziyannis, S.; Hoofnagle, J.H.; Di Bisceglie, A.M.; Gerin, J.L.; Casey, J.L. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology 1997, 234, 160–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dény, P. Hepatitis delta virus genetic variability: From genotypes I, II, III to eight major clades? Curr. Top. Microbiol. Immunol. 2006, 307, 151–171. [Google Scholar] [CrossRef]
- Le Gal, F.; Gault, E.; Ripault, M.-P.; Serpaggi, J.; Trinchet, J.-C.; Gordien, E.; Dény, P. Eighth major clade for hepatitis delta virus. Emerg. Infect. Dis. 2006, 12, 1447–1450. [Google Scholar] [CrossRef] [PubMed]
- Delfino, C.M.; Cerrudo, C.S.; Biglione, M.; Oubiña, J.R.; Ghiringhelli, P.D.; Mathet, V.L. A comprehensive bioinformatics analysis of hepatitis D virus full-length genomes. J. Viral. Hepat. 2018, 25, 860–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wille, M.; Netter, H.J.; Littlejohn, M.; Yuen, L.; Shi, M.; Eden, J.-S.; Klaassen, M.; Holmes, E.C.; Hurt, A.C. A divergent hepatitis D-like agent in birds. Viruses 2018, 19, 720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.-S.; Pattersson, J.H.; Le Lay, C.; Shi, M.; Lo, N.; Wille, M.; Eden, J.-S.; Holmes, E.C. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019, 5, vez021. [Google Scholar] [CrossRef]
- Wang, K.S.; Choo, Q.L.; Weiner, A.J.; Ou, J.H.; Najarian, R.C.; Thayer, R.M.; Mullenbach, G.T.; Denniston, K.J.; Gerin, J.L.; Houghton, M. Structure, sequence, and expression of the hepatitis delta (δ) viral genome. Nature 1986, 323, 508–514. [Google Scholar] [CrossRef]
- Taylor, J.M. The structure and replication of hepatitis delta virus. Annu. Rev. Microbiol. 1992, 46, 253–276. [Google Scholar] [CrossRef]
- Elena, S.F.; Dopazo, J.; Flores, R.; Diener, T.O.; Moya, A. Phylogeny of viroids, viroidlike satellite RNAs, and viroidlike domain of hepatitis d virus RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 5631–5634. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, G.M.; Woelk, C.H.; Rambaut, A.; Holmes, E.C. Testing the extent of sequence similarity among viroids, satellite RNAs, and hepatitis delta virus. J. Mol. Evol. 2000, 50, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Sharmeen, L.; Kuo, M.Y.P.; Dinter-Gottlieb, G.; Taylor, J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988, 62, 2674–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferré-D’Amaré, A.R.; Zhou, K.; Doudna, J.A. Crystal structure of a hepatitis delta virus ribozyme. Narture 1998, 395, 567–574. [Google Scholar] [CrossRef]
- Been, M.D. HDV ribozyme. Curr. Top. Microbiol. Immunol. 2006, 307, 67–89. [Google Scholar] [CrossRef]
- Casey, J.L. RNA editing in hepatitis delta virus. Curr. Top. Microbiol. Immunol. 2006, 307, 67–89. [Google Scholar] [CrossRef]
- Kuo, M.Y.P.; Chao, M.; Taylor, J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: Role of delta antigen. J. Virol. 1989, 63, 1945–1950. [Google Scholar] [CrossRef] [Green Version]
- Chao, M.; Hsieh, S.Y.; Taylor, J. Role of two forms of hepatitis delta virus antigen: Evidence for a mechanism of self-limiting genome replication. J. Virol. 1990, 64, 5066–5069. [Google Scholar] [CrossRef] [Green Version]
- Glenn, J.S.; White, J.M. Trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 1991, 65, 2357–2361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, F.L.; Chen, P.L.; Tu, S.J.; Wang, G.J.; Chen, D.S. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 1991, 88, 8490–8494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, W.S.; Bayer, M.; Taylor, J. Assembly of hepatitis delta virus particles. J. Virol. 1992, 66, 2310–2315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imazeki, F.; Omata, M.; Ohto, M. Heterogeneity and evolution rates of delta virus RNA sequences. J. Virol. 1990, 64, 5594–5599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-M.; Bih, F.-Y.; Chao, Y.-C.; Govindarajan, S.; Lai, M.M.C. Evolution of hepatitis delta virus RNA during chronic infection. Virology 1992, 188, 265–273. [Google Scholar] [CrossRef]
- Chao, Y.-C.; Tang, H.-S.; Hsu, C.-T. Evolution rate of hepatitis delta virus RNA isolated in Taiwan. J. Med. Virol. 1994, 43, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Krushkal, J.; Li, W.-H. Substitution rates in hepatitis D virus. J. Mol. Evol. 1995, 41, 721–726. [Google Scholar] [CrossRef]
- Wu, J.C.; Chiang, T.Y.; Shiue, W.K.; Wang, S.Y.; Sheen, I.J.; Huang, Y.H.; Syu, W.J. Recombination of hepatitis D virus RNA sequences and its implications. Mol. Biol. Evol. 1999, 16, 1622–1632. [Google Scholar] [CrossRef]
- Anisimova, M.; Yang, Z. Molecular evolution of the hepatitis delta virus gene: Recombination or positive selection. J. Mol. Evol. 2004, 59, 815–826. [Google Scholar] [CrossRef] [Green Version]
- Bishal, A.K.; Mukherjee, R.; Chakraborty, C. Synonymous codon usage pattern analysis of hepatitis D virus. Virus Res. 2013, 173, 350–353. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Toh, H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform. 2008, 9, 212. [Google Scholar] [CrossRef] [Green Version]
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990, 28, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L.; Fekete, M.; Stadler, P.F. Secondary structreu prediction for aligned RNA sequences. J. Mol.Biol. 2002, 319, 1059–1066. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Imazeki, F.; Omata, M.; Ohto, M. Complete nucleotide sequence of hepatitis delta virus RNA in Japan. Nucl. Acid. Res. 1991, 19, 5439–5440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, J.L.; Brown, T.L.; Colan, E.J.; Wignall, F.S.; Gerin, J.L. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 1993, 90, 9016–9020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism, 3rd ed.; Munro, H.N., Ed.; Academic Press: Cambridge, MA, USA, 1969; pp. 121–132. [Google Scholar]
- Korber, B. HIV Signature and Sequence Variation Analysis. In Computational and Evolutionary Analysis of HIV Molecular Sequences; Rodrigo, A.G., Learn, G.H., Jr., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 55–72. [Google Scholar]
- Miyata, T.; Yasunaga, T. Molecular evolution of mRNA: A method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 1980, 16, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical; Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 15 February 2021).
- Usman, Z.; Velkov, S.; Prozer, U.; Roggendorf, M.; Frishman, D.; Karimzadeh, H. HDVdb: A comprehensive hepatitis D virus database. Viruses 2020, 12, 538. [Google Scholar] [CrossRef] [PubMed]

| Genotype I (170 Sequences) |
| HQ005371, MN984413, MN984411, MN984408, MN984443, MN984429, KJ744224, MN984407, HQ005367, MN984422, MN984459, MN984449, MN984415, MN984453, MN984452, MN984455, MN984451, MN984450, MN984454, MN984427, MN984437, MN984424, MN984428, MN984457, MN984448, MN984425, MN984447, MN984421, MN984430, MN984423, MN984461, MN984414, MN984432, MN984410, MN984431, MT583796, MN984412, MN984460, MN984416, KJ744217, KJ744216, KJ744215, KJ744214, MN984435, MN984436, MN984409, MN984445, MN984419, MN984441, AB118848, MN984440, MN984438, MN984442, MN984417, MN984439, MN984420, MN984466, MN984458, MH457143, X85253, MT583812, KJ744223, KJ744220, KJ744222, KJ744221, KT722840, MN984456, KF660602, AB118849, MN984426, AY648957, AY648956, KF660600, MN984444, MN984434, AY648959, AF425644, AY648958, AF104263, MH791030, MH791028, KY463681, U81989, MG926381, MK890226, MK890225, MK890231, HQ005372, MG926380, HQ005366, MN984469, MH791029, MK890228, MK890227, MK890232, MK890235, KJ744237, MK890234, HQ005370, HQ005364, MK890230, HQ005368, HQ005365, U81988, AY633627, KJ744255, MN984418, KJ744238, MH791027, MT583805, MT583804, MH457145, KJ744243, MN984463, MN984433, KJ744257, KJ744244, EF514905, EF514907, EF514904, EF514903, EF514906, KJ744256, MN984465, KJ744228, KJ744227, KJ744234, KJ744254, KJ744248, KJ744232, KJ744253, KJ744247, MN984462, KJ744245, MN984464, KJ744218, KJ744249, KJ744231, MN984446, KJ744226, KJ744225, NC_001653, D01075, KJ744230, KJ744229, KJ744235, KJ744233, KJ744250, MK124579, M21012, HM046802, AJ307077, AJ000558, HQ005369, KJ744242, KJ744240, KJ744241, MK890224, MK890229, MG711778, MK890233, KM110794, KM110792, KM110797, KM110799, MG711717, KM110795, KM110791, KM110798, KM110790 |
| Genotype II (51 sequences) |
| KF660599, KF660598, MG557658, AB118846, AY261457, AF425645, AF104264, MK234591, MK234593, MK234592, MK234594, MG557659, MN984468, KM110805, MN984470, MG711777, AB118844, AB118842, AF209859, MT050453, AB118843, AY648954, AY648953, AY648952, AB118847, AY648955, AB118841, MN401236, AB118845, AB118826, AB118835, AB118834, AB118819, AB118828, AB118832, AB118840, AB118830, AB118827, AB118820, AB118825, AB118821, AB118839, AB118822, AB118818, AB118833, AB118838, AB118823, AB118836, AB118824, AB118837, AB118831 |
| Genotype III (5 sequences) |
| AB037947, KC590319, HF679406, HF679405, HF679404 |
| Threshold | Average Ratio |
|---|---|
| 0.4 | 0.753 |
| 0.5 | 0.722 |
| 0.6 | 0.672 |
| 0.7 | 0.623 |
| edbp(x,y)/ednbp(x, y) | edbp(x,y)/ed(x, y) | ednbp(x,y)/ed(x, y) | edbp(x,y)/ | ednbp(x,y)/ | ed(x,y)/ | edbp(x,y)/ | ednbp(x,y)/ | ed(x,y)/ | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | median | 0.46 | 0.59 | 1.3 | 0.16 | 0.34 | 0.27 | 0.52 | 1.1 | 0.89 | 0.30 |
| n = 14,276 | p-value | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 | <2.2 × 10−15 |
| II | median | 0.70 | 0.71 | 1.0 | 0.30 | 0.44 | 0.43 | 0.87 | 1.2 | 1.2 | 0.35 |
| n = 1263 | p-value | <2.2 × 10−15 | <2.2 × 10−15 | 8.6 × 10−4 * | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 * | <2.2 × 10−15 |
| III | median | 0.29 | 0.50 | 1.3 | 0.40 | 0.74 | 0.63 | 0.63 | 1.8 | 1.1 | 0.69 |
| n = 9 | p-value | 1.9 × 10−2 | 1.9 × 10−2 | 1.4 × 10−1 * | 2.0 × 10−2 | 1.0 * | 1.0 * | 2.0 × 10−2 | 2.0 × 10−1 * | 6.4 × 10−1 * | 1.0 |
| edbp(x,y)/ednbp(x, y) | edbp(x,y)/ed(x, y) | ednbp(x,y)/ed(x, y) | edbp(x,y)/ | ednbp(x,y)/ | ed(x,y)/ | edbp(x,y)/ | ednbp(x,y)/ | ed(x,y)/ | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I (vs.) II | median | 0.76 | 0.74 | 0.99 | 0.21 | 0.28 | 0.28 | 0.70 | 0.89 | 0.93 | 0.31 |
| n = 8670 | p-value | <2.2 × 10−15 | <2.2 × 10−15 | <1.8 × 10−7 * | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 |
| I (vs.) III | median | 0.60 | 0.71 | 1.2 | 0.26 | 0.43 | 0.36 | 0.87 | 1.5 | 1.2 | 0.29 |
| n = 850 | p-value | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 * | <2.2 × 10−15 |
| II (vs.) III | median | 0.61 | 0.72 | 1.2 | 0.32 | 0.48 | 0.43 | 0.82 | 1.3 | 1.1 | 0.38 |
| n = 255 | p-value | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 | <2.2 × 10−15 * | <2.2 × 10−15 * | <2.2 × 10−15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagata, S.; Kiyohara, R.; Toh, H. Constraint of Base Pairing on HDV Genome Evolution. Viruses 2021, 13, 2350. https://doi.org/10.3390/v13122350
Nagata S, Kiyohara R, Toh H. Constraint of Base Pairing on HDV Genome Evolution. Viruses. 2021; 13(12):2350. https://doi.org/10.3390/v13122350
Chicago/Turabian StyleNagata, Saki, Ryoji Kiyohara, and Hiroyuki Toh. 2021. "Constraint of Base Pairing on HDV Genome Evolution" Viruses 13, no. 12: 2350. https://doi.org/10.3390/v13122350
APA StyleNagata, S., Kiyohara, R., & Toh, H. (2021). Constraint of Base Pairing on HDV Genome Evolution. Viruses, 13(12), 2350. https://doi.org/10.3390/v13122350





