Retrospective Characterization of Initial Peste des petits ruminants Outbreaks (2008–2012) in the Democratic Republic of the Congo
Abstract
:1. Introduction
2. Materials and Methods
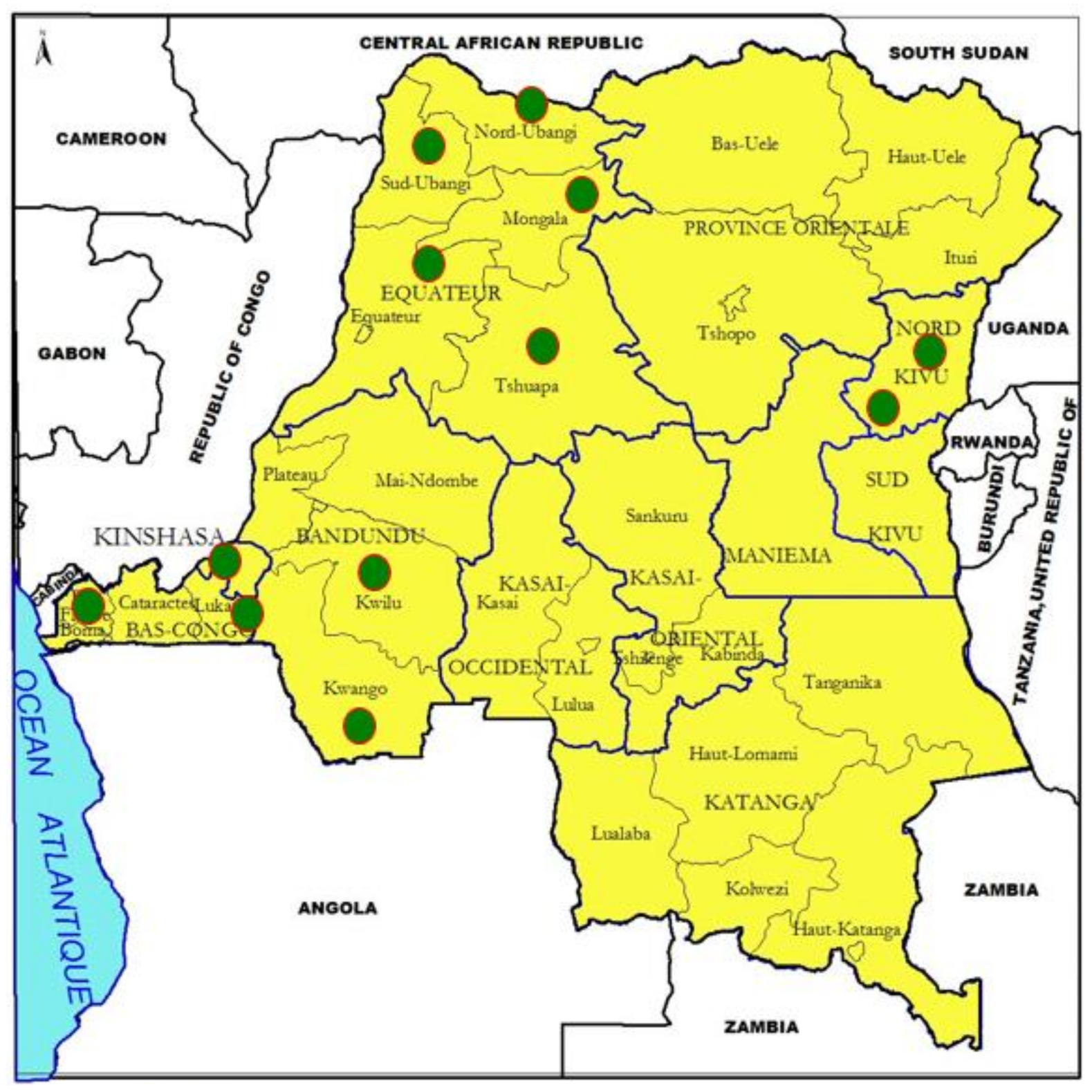
2.1. Study Areas
2.2. Ethical Statement
2.3. Sampling
2.4. Clinical and Postmortem Examination
2.5. Antibody/Antigen Detection
2.6. RNA Extraction, Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Sequencing
2.7. Sequence Datasets
2.8. Risk Factor Assessment
2.9. Estimation of Mortalities
3. Results
3.1. Affected Areas and PPR Clinical Profile
3.2. Antibody Detection
3.3. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
3.4. Sequencing Outputs
3.5. Risk Factor Assessment
3.6. Estimate of Mortalities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahapatra, M.; Sayalel, K.; Muniraju, M.; Eblate, E.; Fyumagwa, R.; Shilinde, S.; MaulidMdaki, M.; Keyyu, J.; Parida, S.; Kock, R. Spillover of Peste des Petits Ruminants Virus from Domestic to Wild Ruminants in the Serengeti Ecosystem, Tanzania. Emerg. Infect. Dis. 2015, 21, 2230–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parida, S.; Muniraju, M.; Mahapatra, M.; Muthuchelvan, D.; Buczkowski, H.; Banyard, A. Peste des petits ruminants. Veter- Microbiol. 2015, 181, 90–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Maherchandani, S.; Kashyap, S.K.; Singh, S.V.; Sharma, S.; Chaubey, K.K.; Ly, H. Peste Des Petits Ruminants Virus Infection of Small Ruminants: A Comprehensive Review. Viruses 2014, 6, 2287–2327. [Google Scholar] [CrossRef] [Green Version]
- Chazya, R.; Muma, J.B.; Mwacalimba, K.; Karimuribo, E.; Mkandawire, E.; Simuunza, M. A Qualitative Assessment of the Risk of Introducing Peste des Petits Ruminants into Northern Zambia from Tanzania. Veter. Med. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- OIE; FAO. Control et Eradication de la Peste des Petits Ruminants: Investir dans les Systèmes Vétérinaires, la Sécurité Ali-Mentaire et la Réduction de la Pauvreté; OIE et FAO 2015 Report; FAO: Italy, Rome, 2015. [Google Scholar]
- Tendonkeng, F.; Pamo, T.E.; Boukila, B.; Defang, T.E.; Njiki, W.; Miégoué, E.; Fogang, Z.B.; Lemoufouet, J.; Djiomika, T.J. Caractéristiques socio-économiques et techniques de l’élevage des petits ruminants dans la région du Cameroun, cas du Département de la Mvila. Livest. Rural Dev. 2013, 25. Available online: http://www.lrrd.org/lrrd25/4/fern25064.htm (accessed on 27 July 2021).
- Woma, T.Y.; Ekong, P.S.; Bwala, D.G.; Ibu, J.O.; Ta’ama, L.; Dyek, D.Y.; Saleh, L.; Shamaki, D.; Kalla, D.J.U.; Bailey, D.; et al. Serosurvey of peste des petits ruminants virus in small ruminants from different agro-ecological zones of Nigeria. Onderstepoort J. Vet. Res. 2016, 83, 1035. [Google Scholar] [CrossRef] [PubMed]
- Bouchemla, F.; Agoltsov, V.A.; Popova, O.M.; Padilo, L.P. Assessment of the Peste des petits ruminants World Epi-Zootic Situation and Estimate Its Spreading to Russia. Available online: www.veterinaryworld.org/Vol.11/May−2018/7.pdf (accessed on 21 July 2021).
- Abubakar, M.; Mahapatra, M.; Muniraju, M.; Arshed, M.J.; Khan, E.H.; Banyard, A.C.; Ali, Q.; Parida, S. Serological detection of antibodies to Peste des petits ruminants virus in large Ruminants. Transbound Emerg. Disease 2017, 64, 513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahur, A.; Irshad, H.; Hussain, M.; Ullah, A.; Jahangir, M.; Khan, M.Q.; Farooq, M.S. The epidemiology of peste des petits ruminants in Pakistan. Rev. Sci. Tech. L’OIE 2008, 27, 877–884. [Google Scholar] [CrossRef]
- FAO. FAO responds to PPR epidemic in the Democraticv Republic of Congo. Vet. Rec. 2012, 171, 7. [Google Scholar] [CrossRef] [PubMed]
- Britton, A.; Caron, A.; Bedane, B. Progress to Control and Eradication of Peste des Petits Ruminants in the Southern African Development Community Region. Front. Veter. Sci. 2019, 6, 343. [Google Scholar] [CrossRef]
- Tshilenge, M.G.; Walandila, J.S.; Byakya, D.; Masumu, J.; Katshay, L.; Cattoli, G.; Bushu, E.; Mpiana, S.; Dundon, W.G. Peste des petitts ruminants viruses of lineages II and III identified in the Democratic Republic of the Congo. Vet. Microbiol. 2019, 239, 108493. [Google Scholar] [CrossRef]
- Radostis, O.; Gay, C.C.; Blood, D.C.; Kenneth, W.H. Peste des petits ruminants. In Veterinary Medecine, a Textbook of the Diseases of Cattle, Sheep, Pigs, Goats and Horses, 9th ed.; W.B. Saunders Company Ltd.: London, UK, 2000; pp. 1077–1079. ISBN 0702026042. [Google Scholar]
- Libeau, G.; Prehaud, C.; Lancelot, R.; Colas, F.; Guerre, L.; Bishop, D.H.L.; Diallo, A. Development of a competitive ELISA for detecting antibodies to the peste des petits ruminants virus using a recombinant nucleoprotein. Res. Vet. Sci. 1995, 58, 50–55. [Google Scholar] [CrossRef]
- Mapaco, L.; Monjane, I.; Fafetine, J.; Arone, D.; Caron, A.; Chilundo, A.; Quembo, C.; Carrilho, M.D.C.; Nhabomba, V.; Zohari, S.; et al. Peste des Petits Ruminants Virus Surveillance in Domestic Small Ruminants, Mozambique (2015 and 2017). Front. Veter. Sci. 2019, 6, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çeribasi, S.; Özkaraca, M.; Çeribasi, A.O.; Özer, H. Immunohistochemical detection of Peste des Petits Ruminants (PPR) Viral antigens in Pneumonic Caprine Lungs in Elazig Region. Available online: http://www.fusabil.org (accessed on 21 July 2021).
- Couacy-Hymann, E.; Roger, F.; Hurard, C.; Guillou, J.; Libeau, G.; Diallo, A. Rapid and sensitive detection of peste des petits ruminants virus by a polymerase chain reaction assay. J. Virol. Methods 2001, 100, 17–25. [Google Scholar] [CrossRef]
- Muniraju, M.; Munir, M.; Parthiban, A.R.; Banyard, A.C.; Bao, J.; Wang, Z.; Ayebazibwe, C.; Ayelet, G.; El Harrak, M.; Mahapatra, M.; et al. Molecular Evolution of Peste des Petits Ruminants Virus. Emerg. Infect. Dis. 2014, 20, 2023–2033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muniraju, M.; Munir, M.; Banyard, A.C.; Ayebazibwe, C.; Wensman, J.; Zohari, S.; Berg, M.; Parthiban, A.R.; Mahapatra, M.; Libeau, G.; et al. Complete Genome Sequences of Lineage III Peste des Petits Ruminants Viruses from the Middle East and East Africa. Genome Announc. 2014, 2, e01023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J.; Wang, Q.; Parida, S.; Liu, C.; Zhang, L.; Zhao, W.; Wang, Z. Complete Genome Sequence of a Peste des Petits Ruminants Virus Recovered from Wild Bharal in Tibet, China. J. Virol. 2012, 86, 10885–10886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Gibson, T.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1, 22. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A.-U.; Abubakar, M.; Rasool, M.H.; Manzoor, S.; Saqalein, M.; Rizwan, M.; Munir, M.; Ali, Q.; Wensman, J.J. Evaluation of Risk Factors for Peste des Petits Ruminants Virus in Sheep and Goats at the Wildlife-Livestock Interface in Punjab Province, Pakistan. BioMed Res. Int. 2016, 2016, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Putt, S.N.H.; Shaw, A.P.M.; Woods, A.J.; Tylor, I.; James, A.D. CGSpace: Montpellier, France, 1987; pp.93−105; ISBN 9290530839.maladies. In Epidémiologie et Economie Vétérinaires en Afrique. Manuel à L’usage des Planifi-Cateurs de la Santé Animale; CGSpace: Montpellier, France, 1987; pp. 93–105. ISBN 9290530839. [Google Scholar]
- Dundon, W.G.; Diallo, A.; Cattoli, G. Peste des petits ruminants in Africa: A review of currently available molecular epide-miological data. Arch. Virol. 2020, 165, 2147–2163. [Google Scholar] [CrossRef]
- Maganga, G.D.; Verrier, D.; Zerbinati, R.M.; Drosten, C.; Drexler, J.F.; Leroy, E.M. Molecular typing of PPRV strains detected during an outbreak in sheep and goats in south-eastern Gabon. Virol. J. 2013, 10, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahapatra, M.; Pachauri, R.; Subramaniam, S.; Banyard, A.C.; ChandraSekar, S.; Ramakrishnan, M.A.; Njeumi, F.; Muthuchelvan, D.; Parida, S. Ongoing Assessment of the Molecular Evolution of Peste Des Petits Ruminants Virus Continues to Question Viral Origins. Viruses 2021, 13, 2144. [Google Scholar] [CrossRef]
- Mantip, S.; Quan, M.; Shamaki, D.; Van Vuuren, M. Comparison of nucleotide sequences of recent and previous lineages of peste-des-petits-ruminants viruses of sheep and goats in Nigeria. Onderstepoort J. Veter. Res. 2016, 83. [Google Scholar] [CrossRef] [PubMed]
- Kitengie, L.J.B.; Mayele, N.B.E. La République Démocratique du Congo en Afrique et Dans le Monde; Imprimeries MEDI-ASPAUL: Kinshasa, Congo, 2013. [Google Scholar]
- Misinzo, G.; Kgotlele, T.; Muse, E.A.; Van Doorsselaere, J.; Berg, M.; Munir, M. Peste des petits ruminants virus lineage II and IV from goats in southern Tanzania during an outbreak. Br. J. Virol. 2014, 2, 1–4. [Google Scholar]
- Kgotlele, T.; Macha, E.S.; Kasanga, C.J.; Kusiluka, L.J.M.; Karimuribo, E.D.; Van Doorsselaere, J.; Wensman, J.J.; Munir, M.; Misinzo, G. Partial Genetic Characterization of Peste des Petits Ruminants Virus from Goats in Northern and Eastern Tanzania. Transbound. Emerg. Dis. 2014, 61, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.A.; Mahapatra, M.; Chubwa, C.; Clarke, B.; Batten, C.; Hicks, H.; Henstock, M.; Keyyu, J.; Kock, R.; Parida, S. Characterisation of Peste des Petits Ruminants Disease in Pastoralist Flocks in Ngorongoro District of Northern Tanzania and Bluetongue Virus Co-Infection. Viruses 2020, 12, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar, X.F.; Mahapatra, M.; Begovoeva, M.; Kalema-Zikusoka, G.; Driciru, M.; Ayebazibwe, C.; Adwok, D.S.; Kock, M.; Lukusa, J.K.; Muro, J.; et al. Peste des Petits Ruminants at the Wildlife-Livestock Interface in the Northern Albertine Rift and Nile Basin, East Africa. Viruses 2020, 12, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| FORMER 11 PROVINCES OF THE DRC | Knowledge Attitude and Practice; public awareness | Easy Contact with other Countries (porous border) | Forest Exploitation (Mining, Agriculture, wood industry and hunting) | Interfaces: Wild, Domestic Animals and humans | Farming System (Backyard with animals scavenging during the day. | Political Crisis with Forced movements of populations including small scale breeders | Poor Biosafety System at the farming level | Rapid Increase in Population with low offer and high demand at the national level | Transhumance and Nomadism (East and North East chiefly) | Climate Change Drought and/Floods; seasons | Lack of Riposte in Real Time | Lack of Quarantine Infrastructures at the borders level | Deforestation, Release of Blood sucking vectors | Low Number of Qualified Human resources | Commercial Exchanges through borders markets (cases of Zongo, Lufu, Pweto, Cabinda) | Lack of Systematic Vaccination campaigns programme | Lack of Surveillance and Fences in wildlife reserves (cases of Virunga, Caramba, etc.) | Poor Capacity of Communication and reporting. including for alerts, | Risk Cartography |
| Equateur | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 17 |
| Bandundu | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 14 |
| Bas Congo | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 15 |
| Kasaï Or. | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 12 |
| Kasaï Occ. | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 |
| Katanga | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 13 |
| Kinshasa | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 10 |
| Maniema | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 11 |
| North Kivu | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 14 |
| Orientale | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 18 |
| South Kivu | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 13 |
| Risk weight | 11 | 8 | 5 | 7 | 10 | 4 | 11 | 7 | 2 | 11 | 11 | 11 | 7 | 7 | 8 | 11 | 11 | 11 |
| Country | Lineages of PPRV Outbreaks | Year of Outbreak with Cited References |
|---|---|---|
| Central African Rep (CAR) | IV | 2004 [26] |
| South Sudan | IV | 2011 [26] |
| Uganda | IV, III | 2012 (IV) and 2014 (III) [20,26] |
| Tanzania | II, III, IV | 2008 (seropositive), 2013 (III), 2011 (II and IV) [1,31,32,33] |
| Angola | IV | 2012 [26] |
| Gabon | IV | 2011, 2007 (seropositive) [26,30] |
| Cameroon | IV | 1997, 2017 [26] |
| Burundi | III | 2017 [26] |
| DRC | II, III, IV | 2011−2012 (IV, in this study), 2016−18 (II and III) [13,34] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulumba-Mfumu, L.K.; Mahapatra, M.; Diallo, A.; Clarke, B.; Twabela, A.; Matondo-Lusala, J.P.; Njeumi, F.; Parida, S. Retrospective Characterization of Initial Peste des petits ruminants Outbreaks (2008–2012) in the Democratic Republic of the Congo. Viruses 2021, 13, 2373. https://doi.org/10.3390/v13122373
Mulumba-Mfumu LK, Mahapatra M, Diallo A, Clarke B, Twabela A, Matondo-Lusala JP, Njeumi F, Parida S. Retrospective Characterization of Initial Peste des petits ruminants Outbreaks (2008–2012) in the Democratic Republic of the Congo. Viruses. 2021; 13(12):2373. https://doi.org/10.3390/v13122373
Chicago/Turabian StyleMulumba-Mfumu, Leopold K., Mana Mahapatra, Adama Diallo, Brian Clarke, Augustin Twabela, Jean Pierre Matondo-Lusala, Felix Njeumi, and Satya Parida. 2021. "Retrospective Characterization of Initial Peste des petits ruminants Outbreaks (2008–2012) in the Democratic Republic of the Congo" Viruses 13, no. 12: 2373. https://doi.org/10.3390/v13122373
APA StyleMulumba-Mfumu, L. K., Mahapatra, M., Diallo, A., Clarke, B., Twabela, A., Matondo-Lusala, J. P., Njeumi, F., & Parida, S. (2021). Retrospective Characterization of Initial Peste des petits ruminants Outbreaks (2008–2012) in the Democratic Republic of the Congo. Viruses, 13(12), 2373. https://doi.org/10.3390/v13122373







