Identification of a Novel Geminivirus in Fraxinus rhynchophylla in Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. HTS and Genome Assembly
2.3. Detection of the Novel Virus in F. rhynchophylla
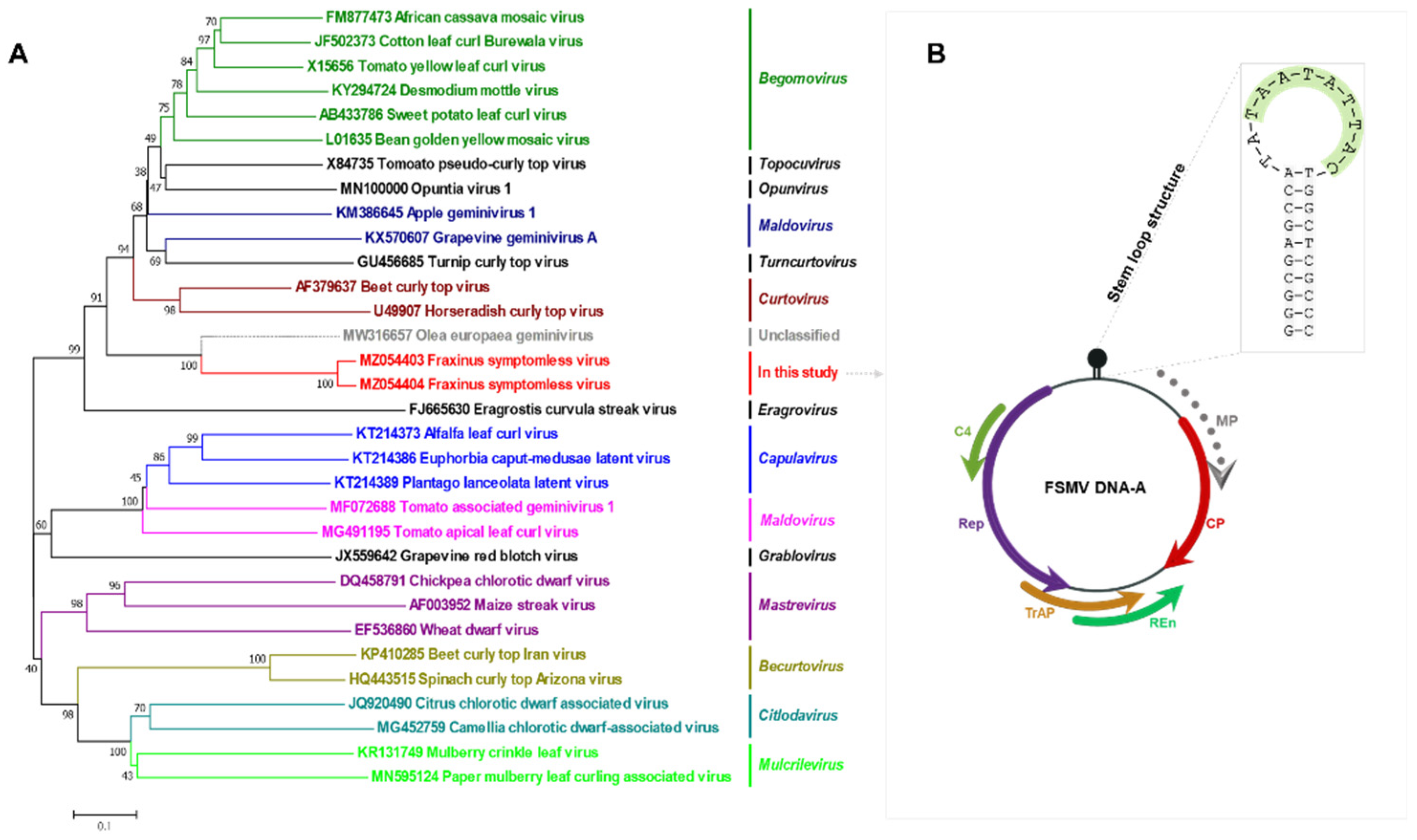
2.4. Genome Organization and Homology Searches for Genes
2.5. Attempts at Further Characterization by Southern Blot Hybridization
2.6. Strand-Specific PCR for Virus Detection
2.7. Construction of Infectious Clone of FSMV
2.8. Agro-Inoculation with the FSMV Infectious Clone
3. Results
3.1. HTS Results
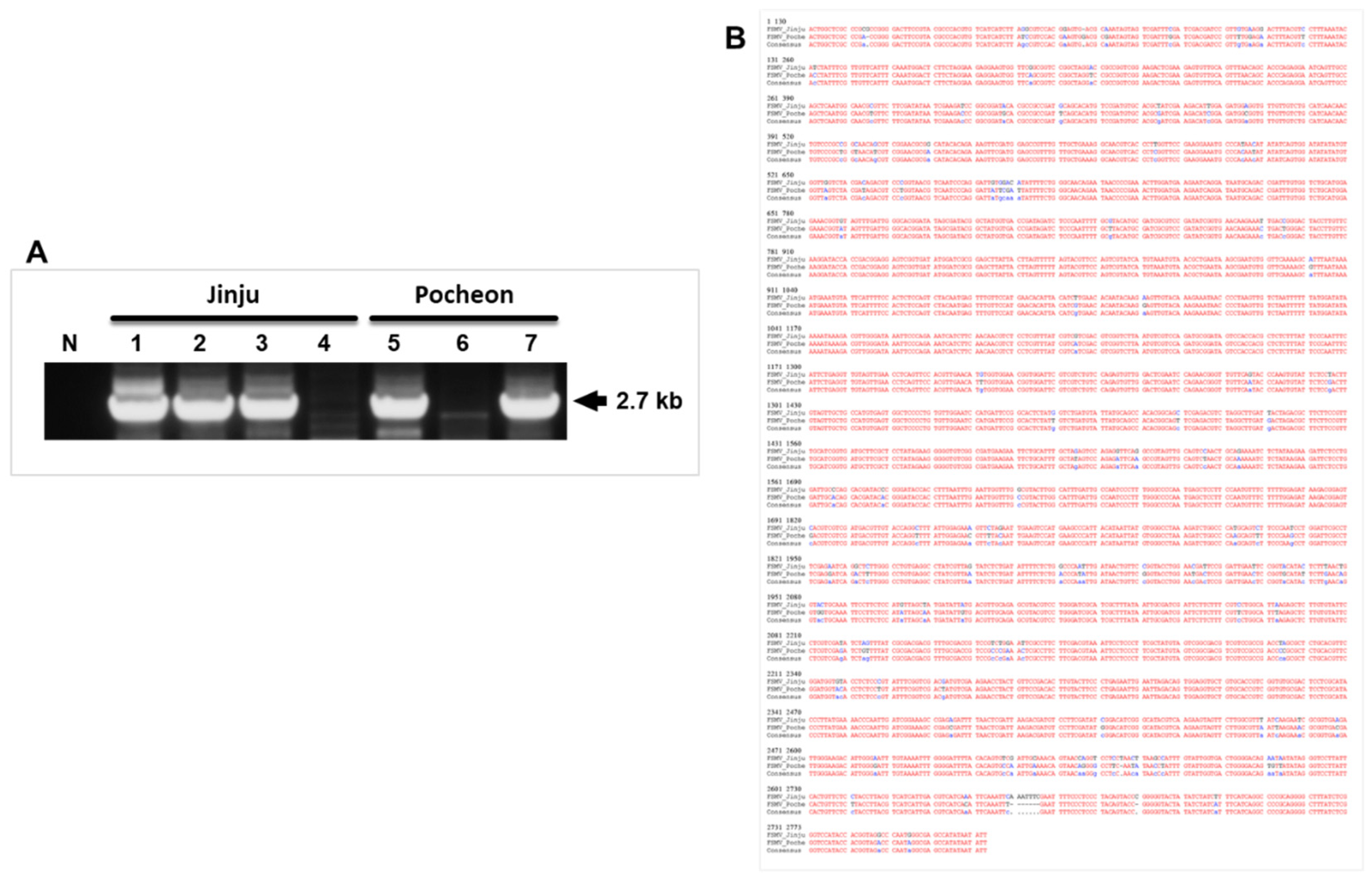
3.2. Virus Detection through PCR
3.3. Genomic Features
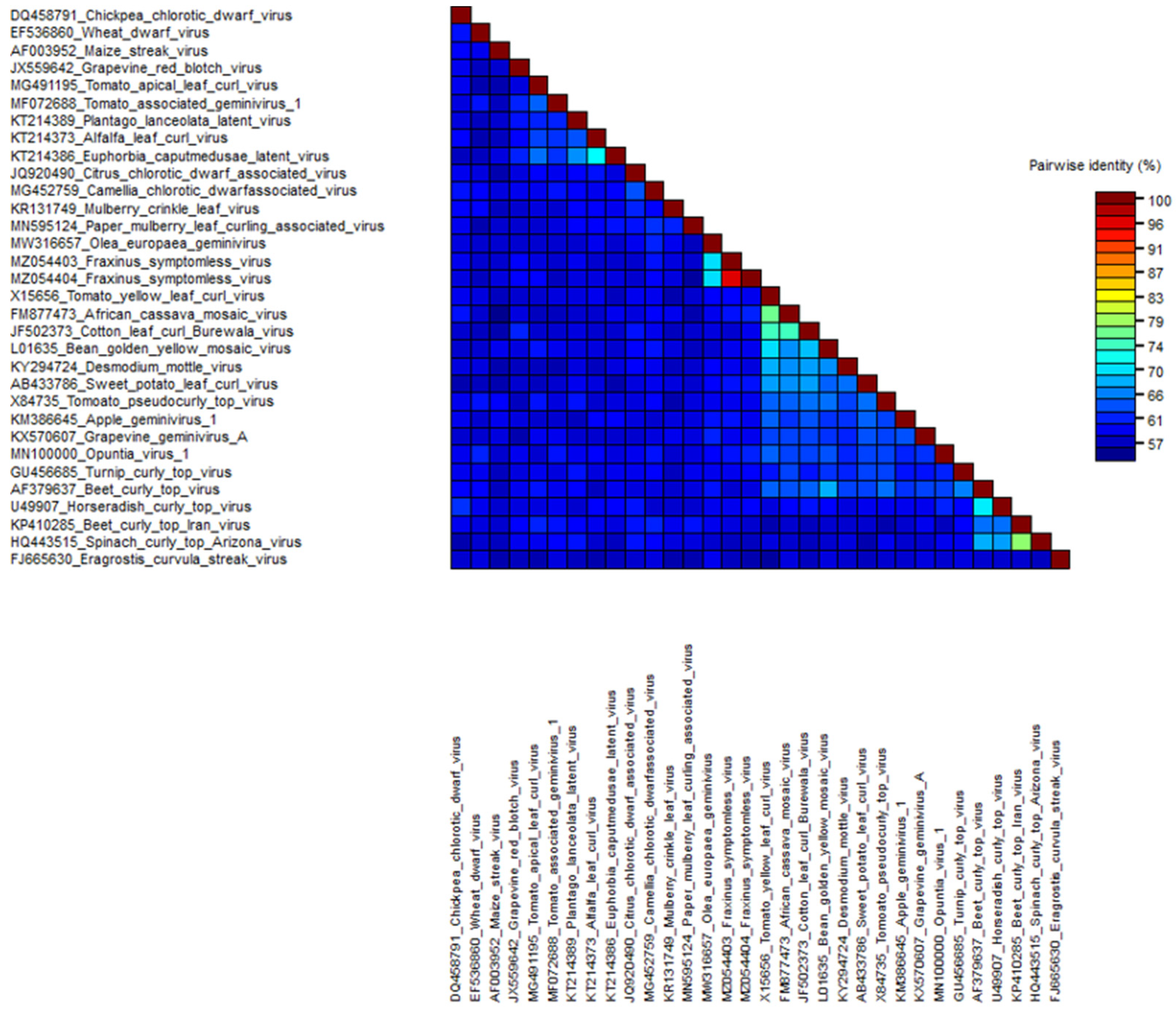
3.4. Phylogenetic Relationship with Other Virus Families
3.5. Southern Blotting Hybridization Analysis
3.6. Strand-Specific PCR and Site-Based Detection
3.7. Infectivity through Infectious Clone Inoculation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sarfraz, I.; Rasul, A.; Jabeen, F.; Younis, T.; Zahoor, M.K.; Arshad, M.; Ali, M. Fraxinus: A Plant with Versatile Pharmacological and Biological Activities. eCAM 2017, 2017, 4269868. [Google Scholar] [CrossRef] [Green Version]
- Younis, T.; Khan, M.R.; Sajid, M.; Majid, M.; Zahra, Z.; Shah, N.A. Fraxinus xanthoxyloides leaves reduced the level of inflammatory mediators during in vitro and in vivo studies. BMC Complement Altern. Med. 2016, 16, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Manjarrés, J.; Gérard, P.; Dufour, J.; Raquin, C. Differential patterns of morphological and molecular hybridization between Fraxinus excelsior L. and Fraxinus angustifolia Vahl (Oleaceae) in east ern and western France. Mol. Ecol. 2006, 15, 3245–3257. [Google Scholar] [CrossRef]
- Hinsinger, D.D.; Gaudeul, M.; Couloux, A.; Bousquet, J.; Frascaria-Lacoste, N. The phylogeography of Eurasian Fraxinus species reveals ancient transcontinental reticulation. Mol. Phylogenet. Evol. 2014, 77, 223–237. [Google Scholar] [CrossRef]
- Kostova, I.; Iossifova, T. Chemical components of Fraxinus species. Fitoterapia 2007, 78, 85–106. [Google Scholar] [CrossRef]
- Wu, Z.-B.; Liu, Y.; Tian, S.-S.; Wen, C. Chemical constituents of the stem bark of Fraxinus rhynchophylla. Chem. Nat. Compd. 2014, 49, 1162–1163. [Google Scholar] [CrossRef]
- Torres, M.; Palomares, O.; Quiralte, J.; Pauli, G.; Rodríguez, R.; Villalba, M. An Enzymatically active β-1, 3-Glucanase from ash pollen with allergenic properties: A particular member in the Oleaceae Family. PLoS ONE 2015, 10, e0133066. [Google Scholar]
- Jung, B.-N.; Choi, Y.-J.; Shin, H.-D.; Park, J.-H. Macruropyxis fraxini on Fraxinus rhynchophylla: Confirmation in the Korean Peninsula after 82 Years and the First Record in South Korea. Mycobiology 2020, 48, 518–521. [Google Scholar] [CrossRef]
- Luo, Y.; Cobb, R.E.; Zhao, H. Recent advances in natural product discovery. Curr. Opin. Biotechnol. 2014, 30, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassanis, B.; Macfarlane, I. Transmission of Tobacco Necrosis Virus to Tobacco Callus Tissues by Zoospores of Olpidium brassicae. Nature 1964, 201, 218–219. [Google Scholar] [CrossRef]
- Reichmann, M. The satellite tobacco necrosis virus: A single protein and its genetic code. Proc. Natl. Acad. Sci. USA 1964, 52, 1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, I. Characteristics of a satellite-like virus of tobacco ringspot virus. Virol. J. 1971, 45, 108–122. [Google Scholar] [CrossRef]
- Hollings, M.; Stone, O.; Dale, W. Tomato ringspot virus in Pelargonium in England. Plant Pathol. 1972, 21, 46–47. [Google Scholar]
- Thomas, W.; Procter, C. Arabis mosaic virus in Cyphomandra betaceae Sendt. N. Z. J. Agric. Res. 1972, 15, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.M.A. Cotranslational disassembly of tobacco mosaic virus in vitro. Virol. J. 1984, 137, 255–265. [Google Scholar] [CrossRef]
- Kay, L.E.W.M. Stanley’s crystallization of the tobacco mosaic virus, 1930–1940. Isis Int. Rev. Devoted Hist. Sci. Its Cult. Influ. 1986, 77, 450–472. [Google Scholar] [CrossRef]
- Harrison, B.D.; Murant, A.F. Nepoviruses: Ecology and Control. In The Plant Viruses: Polyhedral Virions and Bipartite RNA Genomes; Harrison, B.D., Murant, A.F., Eds.; Springer US: Boston, MA, USA, 1996; pp. 211–228. [Google Scholar]
- Quadt-Hallmann, A.; Löw, A.; Hamacher, J. Distribution of cherry leaf roll nepovirus (CLRV) in leaves of deciduous forest trees and herbaceous plants detected by tissue print immunopressblotting (TPI) of whole leaf blades/Verteilung des Kirschenblattroll-Nepovirus (CLRV) in Blättern von Forstgehölzen und krautigen Pflanzen. Virusnachweis an ganzen Blattspreiten mit dem Immungewebepressabdruckverfahren. J. Plant Dis. Prot. 1996, 103, 449–454. [Google Scholar]
- Fillhart, R.C.; Bachand, G.D.; Castello, J.D. Detection of Infectious Tobamoviruses in Forest Soils. Appl. Environ. Microbiol. 1998, 64, 1430–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High Throughput Sequencing For Plant Virus Detection and Discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Negrete, E.A.; Morales-Aguilar, J.J.; Domínguez-Duran, G.; Torres-Devora, G.; Camacho-Beltrán, E.; Leyva-López, N.E.; Voloudakis, A.E.; Bejarano, E.R.; Méndez-Lozano, J. High-Throughput Sequencing Reveals Differential Begomovirus Species Diversity in Non-Cultivated Plants in Northern-Pacific Mexico. Viruses 2019, 11, 594. [Google Scholar]
- Fontenele, R.S.; Salywon, A.M.; Majure, L.C.; Cobb, I.N.; Bhaskara, A.; Avalos-Calleros, J.A.; Argüello-Astorga, G.R.; Schmidlin, K.; Khalifeh, A.; Smith, K.; et al. A Novel Divergent Geminivirus Identified in Asymptomatic New World Cactaceae Plants. Viruses 2020, 12, 398. [Google Scholar] [CrossRef] [Green Version]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; Consortium, I.R. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [Green Version]
- Perry, K.L.; McLane, H.; Thompson, J.R.; Fuchs, M. A novel grablovirus from non-cultivated grapevine (Vitis sp.) in North America. Arch. Virol. 2018, 163, 259–262. [Google Scholar] [CrossRef]
- Loconsole, G.; Saldarelli, P.; Doddapaneni, H.; Savino, V.; Martelli, G.P.; Saponari, M. Identification of a single-stranded DNA virus associated with citrus chlorotic dwarf disease, a new member in the family Geminiviridae. Virol. J. 2012, 432, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Navarro, B.; Zhang, Z.; Lu, M.; Zhou, X.; Chi, S.; Di Serio, F.; Li, S. Identification and molecular characterization of a novel monopartite geminivirus associated with mulberry mosaic dwarf disease. J. Gen. Virol. 2015, 96, 2421–2434. [Google Scholar] [PubMed]
- Liang, P.; Navarro, B.; Zhang, Z.; Wang, H.; Lu, M.; Xiao, H.; Wu, Q.; Zhou, X.; Di Serio, F.; Li, S. Identification and characterization of a novel geminivirus with a monopartite genome infecting apple trees. J. Gen. Virol. 2015, 96, 2411–2420. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, P.; Li, M.; Tian, X.; Zhou, C.; Cao, M. Discovery of a novel geminivirus associated with camellia chlorotic dwarf disease. Arch. Virol. 2018, 163, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Vaghi Medina, C.G.; Teppa, E.; Bornancini, V.A.; Flores, C.R.; Marino-Buslje, C.; López Lambertini, P.M. Tomato Apical Leaf Curl Virus: A Novel, Monopartite Geminivirus Detected in Tomatoes in Argentina. Front. Microbiol. 2018, 8, 2214. [Google Scholar] [CrossRef] [PubMed]
- Fontenele, R.S.; Abreu, R.A.; Lamas, N.S.; Alves-Freitas, D.M.T.; Vidal, A.H.; Poppiel, R.R.; Melo, F.L.; Lacorte, C.; Martin, D.P.; Campos, M.A.; et al. Passion Fruit Chlorotic Mottle Virus: Molecular Characterization of a New Divergent Geminivirus in Brazil. Viruses 2018, 10, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontenele, R.S.; Lamas, N.S.; Lacorte, C.; Lacerda, A.L.M.; Varsani, A.; Ribeiro, S.G. A novel geminivirus identified in tomato and cleome plants sampled in Brazil. Virus Res. 2017, 240, 175–179. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, X.; Jiang, X.; Wu, X.; Luan, X.; Cheng, X. Molecular characterization of common bean curly stunt virus: A novel recombinant geminivirus in China. Arch. Virol. 2020, 165, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Briddon, R.W.; Zafar, Y.; Stanley, J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003, 8, 128–134. [Google Scholar] [CrossRef]
- Jeske, H. Geminiviruses. In TT Viruses: The Still Elusive Human Pathogens; de Villiers, E.-M., Hausen, H.Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 185–226. [Google Scholar]
- Lazarowitz, S.G.; Shepherd, R.J. Gemi-iviruses: Genome structure and gene function. Crit. Rev. Plant Sci 1992, 11, 327–349. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for Plant DNA Replication, Transcription, and Cell Cycle Regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Hehnle, S.; Wege, C.; Jeske, H. Interaction of DNA with the movement proteins of geminiviruses revisited. Virol. J. 2004, 78, 7698–7706. [Google Scholar] [CrossRef] [Green Version]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Bustamante, R.F.R.; Zerbini, F.M.; Adkins, S.; et al. World Management of Geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Moffat, A.S. Geminiviruses Emerge as Serious Crop Threat. Science 1999, 286, 1835. [Google Scholar] [CrossRef]
- Lal, A.; Vo, T.T.B.; Sanjaya, I.G.N.P.W.; Ho, P.T.; Kim, J.-K.; Kil, E.-J.; Lee, S. Nanovirus Disease Complexes: An Emerging Threat in the Modern Era. Front. Plant Sci. 2020, 11, 1829. [Google Scholar] [CrossRef]
- Kraberger, S.; Geering, A.D.W.; Walters, M.; Martin, D.P.; Varsani, A. Novel mastreviruses identified in Australian wild rice. Virus Res. 2017, 238, 193–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiumenti, M.; Greco, C.; De Stradis, A.; Loconsole, G.; Cavalieri, V.; Altamura, G.; Zicca, S.; Saldarelli, P.; Saponari, M. Olea Europaea Geminivirus: A Novel Bipartite Geminivirid Infecting Olive Trees. Viruses 2021, 13, 481. [Google Scholar] [CrossRef]
- Lal, A.; Kil, E.-J.; Vo, T.T.B.; Fadhila, C.; Ho, P.T.; Shuja, M.N.; Ali, M.; Lee, S. First Report of Duranta leaf curl virus Infecting Ficus virens Showing Leaf Curl Symptoms in Pakistan. Plant Dis. 2020, 104, 2034. [Google Scholar] [CrossRef] [Green Version]
- Lal, A.; Kil, E.-J.; Rauf, K.; Ali, M.; Lee, S. First Report of Papaya leaf curl virus Associated with Leaf Curl Disease in Cestrum nocturnum in Pakistan. Plant Dis. 2020, 104, 3089. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Coutts, B.A. Spread of introduced viruses to new plants in natural ecosystems and the threat this poses to plant biodiversity. Mol. Plant Pathol. 2015, 16, 541–545. [Google Scholar] [CrossRef] [Green Version]
- Alexander, H.M.; Mauck, K.E.; Whitfield, A.E.; Garrett, K.A.; Malmstrom, C.M. Plant-virus interactions and the agro-ecological interface. Eur. J. Plant Pathol. 2014, 138, 529–547. [Google Scholar] [CrossRef]
- Elena, S.F.; Fraile, A.; García-Arenal, F. Evolution and Emergence of Plant Viruses. In Advance Virus Research; Maramorosch, K., Murphy, F.A., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 88, pp. 161–191. [Google Scholar]
- Jones, R.A.C. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009, 141, 113–130. [Google Scholar] [CrossRef]
- Shepherd, L.D.; McLay, T.G. Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. J. Plant Res. 2011, 124, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, D.N.; Martin, D.P.; Lefeuvre, P.; Monjane, A.L.; Owor, B.E.; Rybicki, E.P.; Varsani, A. A protocol for the rapid isolation of full geminivirus genomes from dried plant tissue. J. Virol. Methods 2008, 149, 97–102. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briddon, R.; Bull, S.; Mansoor, S.; Amin, I.; Markham, P. Universal primers for the PCR-mediated amplification of DNA β Mol. Biotechnol. 2002, 20, 315. [Google Scholar]
- Bull, S.; Briddon, R.; Markham, P. Universal primers for the PCR-mediated amplification of DNA 1: A satellite-like molecule associated with begomovirus-DNA β complexes. Mol. Biotechnol. 2003, 23, 83–86. [Google Scholar] [CrossRef]
- Rojas, M.; Gilbertson, R.; Maxwell, D. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 1993, 77, 340–347. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Evolution, MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; et al. Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020). Arch. Virol. 2020, 165, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975, 98, 503–517. [Google Scholar] [CrossRef]
- Kil, E.-J.; Kim, S.; Lee, Y.-J.; Byun, H.-S.; Park, J.; Seo, H.; Kim, C.-S.; Shim, J.-K.; Lee, J.-H.; Kim, J.-K.; et al. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016, 6, 19013. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Negrete, E.A.; Sánchez-Campos, S.; Cañizares, M.C.; Navas-Castillo, J.; Moriones, E.; Bejarano, E.R.; Grande-Pérez, A. A sensitive method for the quantification of virion-sense and complementary-sense DNA strands of circular single-stranded DNA viruses. Sci. Rep. 2014, 4, 6438. [Google Scholar] [CrossRef] [Green Version]
- Kil, E.-J.; Park, J.; Choi, E.-Y.; Byun, H.-S.; Lee, K.-Y.; An, C.G.; Lee, J.-H.; Lee, G.-S.; Choi, H.-S.; Kim, C.-S.; et al. Seed transmission of Tomato yellow leaf curl virus in sweet pepper (Capsicum annuum). Eur. J. Plant Pathol. 2018, 150, 759–764. [Google Scholar] [CrossRef]
- Seol, E.; Jung, Y.; Lee, J.; Cho, C.; Kim, T.; Rhee, Y.; Lee, S. In planta transformation of Notocactus scopa cv. Soonjung by Agrobacterium tumefaciens. Plant Cell Rep. 2008, 27, 1197–1206. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Alabi, O.J.; Westrick, N.M.; Golino, D. Prunus geminivirus A: A Novel Grablovirus Infecting Prunus spp. Plant Dis. 2018, 102, 1246–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Rwahnih, M.; Alabi, O.J.; Westrick, N.M.; Golino, D.; Rowhani, A. Description of a Novel Monopartite Geminivirus and Its Defective Subviral Genome in Grapevine. Phytopathology 2017, 107, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, Weighting, and Phylogenetic Utility of Mitochondrial Gene Sequences and a Compilation of Conserved Polymerase Chain Reaction Primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Wang, J.-F.; Jiang, L.-Y.; Qiao, G.-X. Use of a mitochondrial COI sequence to identify species of the subtribe Aphidina (Hemiptera, Aphididae). Zookeys 2011, 1, 1–17. [Google Scholar]
- Hak, H.; Levy, Y.; Chandran, S.A.; Belausov, E.; Loyter, A.; Lapidot, M.; Gafni, Y. TYLCV-Is movement in planta does not require V2 protein. Virol. J. 2015, 477, 56–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
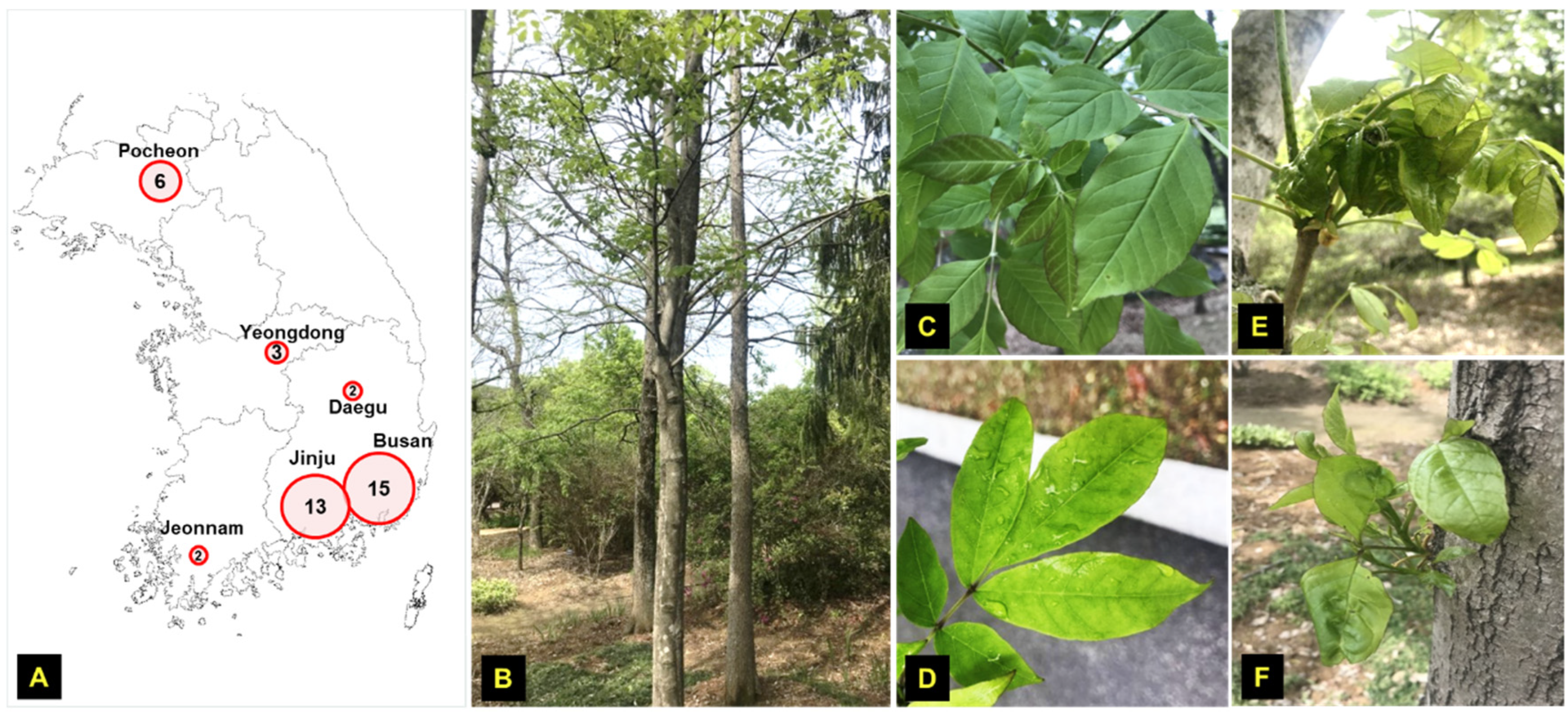




| No. | Location | Collection Date | Labelled as |
|---|---|---|---|
| 1 | Jinju | March 2019 | J1 |
| 2 | J2 | ||
| 3 | J3 | ||
| 4 | J4 | ||
| 5 | September 2019 | J5 | |
| 6 | J6 | ||
| 7 | J7 | ||
| 8 | J8 | ||
| 9 | J9 | ||
| 10 | J10 | ||
| 11 | J11 | ||
| 12 | J12 | ||
| 13 | J13 | ||
| 14 | Busan | October 2019 | B1 |
| 15 | B2 | ||
| 16 | B3 | ||
| 17 | B4 | ||
| 18 | B5 | ||
| 19 | B6 | ||
| 20 | B7 | ||
| 21 | B8 | ||
| 22 | B9 | ||
| 23 | Pocheon | September 2019 | P1 |
| 24 | P2 | ||
| 25 | P3 | ||
| 26 | P4 | ||
| 27 | P5 | ||
| 28 | P6 | ||
| 29 | Jeonnam | September 2019 | JM1 |
| 30 | JM2 | ||
| 31 | Yeongdong | September 2019 | Y1 |
| 32 | Y2 | ||
| 33 | Y3 | ||
| 34 | Daegu | September 2019 | D1 |
| 35 | D2 | ||
| 36 | Busan | May 2020 | B1 * |
| 37 | B2 * | ||
| 38 | B3 * | ||
| 39 | B4 * | ||
| 40 | B5 * | ||
| 41 | B6 * |
| # | ORF | Locus | nt/aa | Strand | Protein |
|---|---|---|---|---|---|
| 1 | V1 | 153–905 | 753/251 | Positive | Coat Protein |
| 2 | V2* | 85–416 | 372/123 | Positive | MP |
| 3 | C3 | 902–1342 | 441/147 | Negative | REn |
| 4 | C2 | 1029–1487 | 459/153 | Negative | TrAP |
| 5 | C1 | 1399–2481 | 1083/361 | Negative | Rep |
| 6 | C4 | 2255–2416 | 162/54 | Negative | C4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lal, A.; Kim, Y.-H.; Vo, T.T.B.; Wira Sanjaya, I.G.N.P.; Ho, P.T.; Byun, H.-S.; Choi, H.-S.; Kil, E.-J.; Lee, S. Identification of a Novel Geminivirus in Fraxinus rhynchophylla in Korea. Viruses 2021, 13, 2385. https://doi.org/10.3390/v13122385
Lal A, Kim Y-H, Vo TTB, Wira Sanjaya IGNP, Ho PT, Byun H-S, Choi H-S, Kil E-J, Lee S. Identification of a Novel Geminivirus in Fraxinus rhynchophylla in Korea. Viruses. 2021; 13(12):2385. https://doi.org/10.3390/v13122385
Chicago/Turabian StyleLal, Aamir, Yong-Ho Kim, Thuy Thi Bich Vo, I Gusti Ngurah Prabu Wira Sanjaya, Phuong Thi Ho, Hee-Seong Byun, Hong-Soo Choi, Eui-Joon Kil, and Sukchan Lee. 2021. "Identification of a Novel Geminivirus in Fraxinus rhynchophylla in Korea" Viruses 13, no. 12: 2385. https://doi.org/10.3390/v13122385
APA StyleLal, A., Kim, Y.-H., Vo, T. T. B., Wira Sanjaya, I. G. N. P., Ho, P. T., Byun, H.-S., Choi, H.-S., Kil, E.-J., & Lee, S. (2021). Identification of a Novel Geminivirus in Fraxinus rhynchophylla in Korea. Viruses, 13(12), 2385. https://doi.org/10.3390/v13122385






