Pathological Features of Echovirus-11-Associated Brain Damage in Mice Based on RNA-Seq Analysis
Abstract
:1. Introduction
2. Methods
2.1. Ethics Statement
2.2. Mice, Cells, and Viruses
2.3. Animal Infection Experiments
2.4. Virus Titration
2.5. Histology of E11-Infected Mice
2.6. Transcriptome Sequencing
2.7. RNA-Seq Data Analysis
2.8. Immunofluorescence Analysis of Brain Tissues
2.9. Bright-Field Microscopy
2.10. Immunofluorescence Analysis of Cells
3. Results
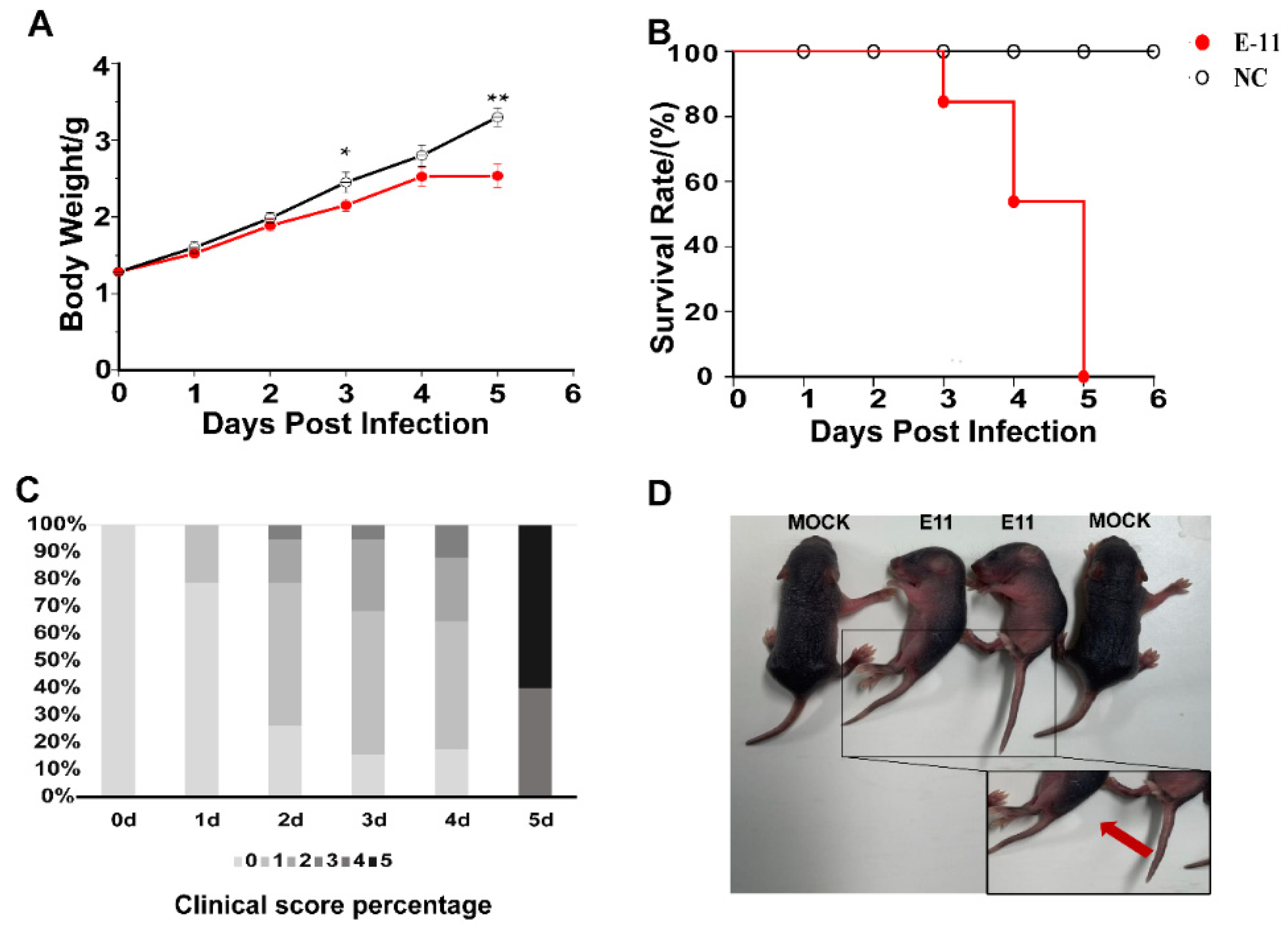
3.1. E11 Challenge in Neonatal IFNAR−/− Mice
3.2. Dynamic Monitoring of Viral Loads in Different Organs
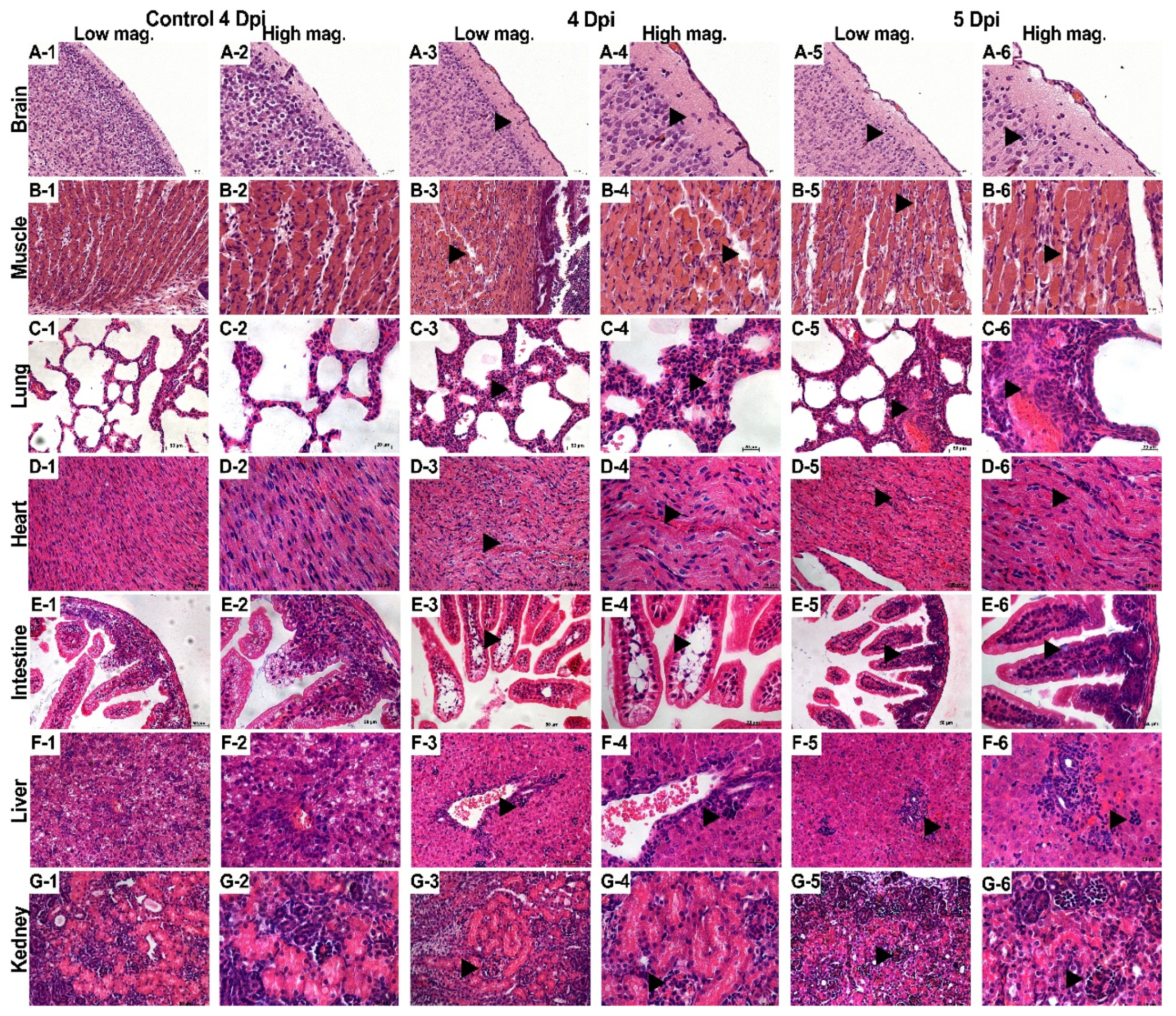
3.3. Pathological Observations
3.4. Transcriptional Changes in E11-Infected Brain and Muscle Tissues
3.5. E11 Replication in the Mouse Brain and in U251 Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitley, R.J.; Gnann, J.W. Viral encephalitis: Familiar infections and emerging pathogens. Lancet 2002, 359, 507–513. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Yang, S.-L.; Yang, H.; Lin, T.-Y.; Hsieh, Y.-C.; Huang, K.-Y.A.; Kuo, C.-Y.; Chiu, C.-H.; Huang, Y.-C.; Chu, S.-M.; et al. Clinical characteristics of echovirus 11 and coxsackievirus B5 infections in Taiwanese children requiring hospitalization. J. Microbiol. Immunol. Infect. 2021, 54, 581–587. [Google Scholar] [CrossRef]
- Lu, J.; Kang, M.; Zeng, H.; Zhong, Y.; Fang, L.; Zheng, X.; Liu, L.; Yi, L.; Lin, H.; Peng, J.; et al. Tracking echovirus eleven outbreaks in Guangdong, China: A metatranscriptomic, phylogenetic, and epidemiological study. Virus Evol. 2020, 6, veaa029. [Google Scholar] [CrossRef]
- Miwa, C.; Sawatari, S. Epidemic of Echo 11 Virus Infection in Gifu Prefecture in 1993. Kansenshogaku Zasshi 1994, 68, 1251–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.R.; Daniel, J.; Mathan, V.I. An epidemic of acute diarrhoea in rural southern India associated with echovirus type 11 infection. Epidemiol. Infect. 1985, 95, 483–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somekh, E.; Shohat, T.; Handsher, R.; Serour, F. An outbreak of echovirus 11 in a children’s home. Epidemiol. Infect. 2001, 126, 441–444. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.-K.; Chai, F.; Li, H.-Y.; Xiao, G.; Guo, L. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J. Proteome Res. 2013, 12, 2666–2678. [Google Scholar] [CrossRef]
- Sun, J.; Ye, F.; Wu, A.; Yang, R.; Pan, M.; Sheng, J.; Jiang, T. Comparative Transcriptome Analysis Reveals the Intensive Early Stage Responses of Host Cells to SARS-CoV-2 Infection. Front. Microbiol. 2020, 11, 593857. [Google Scholar] [CrossRef] [PubMed]
- Van den Broek, M.F.; Müller, U.; Huang, S.; Aguet, M.; Zinkernagel, R.M. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J. Virol. 1995, 69, 4792–4796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, X.; Carr, M.; Zhou, H.; Li, J.; Liu, S.; Liu, T.; Xing, W.; Shi, W. A neonatal murine model of coxsackievirus A4 infection for evaluation of vaccines and antiviral drugs. Emerg. Microbes Infect. 2019, 8, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, H.; Zhang, Y.; Wang, J.; Che, Y.; Dong, C.; Zhang, X.; Na, R.; Shi, H.; Jiang, L.; et al. Neonatal rhesus monkey is a potential animal model for studying pathogenesis of EV71 infection. Virology 2011, 412, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.; Guo, S.; Fan, S.; Zeng, X.; Zhang, Y.; Liao, Y.; Wang, J.; Zhao, T.; Wang, L.; Che, Y.; et al. The Preferential Infection of Astrocytes by Enterovirus 71 Plays a Key Role in the Viral Neurogenic Pathogenesis. Front. Cell. Infect. Microbiol. 2016, 6, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, F.M.S.; Nyo, M.; Wong, A.A.; Tan, N.W.H.; Koh, M.T.; Chan, Y.F.; Chong, C.Y.; Chu, J.J.H. Cytokine and Chemokine Profiling in Patients with Hand, Foot and Mouth Disease in Singapore and Malaysia. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, D.; Sun, T.; Du, Y.; Gao, Y.; Ding, R.; Ji, W.; Zhang, W.; Yang, H.; Chen, S.; et al. Pathological Features of Enterovirus 71-Associated Brain and Lung Damage in Mice Based on Quantitative Proteomic Analysis. Front. Microbiol. 2021, 12, 663019. [Google Scholar] [CrossRef] [PubMed]
- Yin-ping, L.; Bao-ju, W.; Ji-hua, D.; Zhao, L.; Shi-he, G.; Meng-ji, L.; Dong-liang, Y. Antiviral Effect of Interferon-Induced Guanylate Binding Protein-1 against Coxsackie Virus and Hepatitis B Virus B3 in Vitro. Virol. Sin. 2007, 22, 193–198. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.C.-Y.; Taylor, M.W.; Zlotnick, A. In Vitro Assembly of an Empty Picornavirus Capsid follows a Dodecahedral Path. J. Virol. 2012, 86, 13062–13069. [Google Scholar] [CrossRef] [Green Version]
- Koestner, W.; Spanier, J.; Klause, T.; Tegtmeyer, P.-K.; Becker, J.; Herder, V.; Borst, K.; Todt, D.; Lienenklaus, S.; Gerhauser, I.; et al. Interferon-beta expression and type I interferon receptor signaling of hepatocytes prevent hepatic necrosis and virus dissemination in Coxsackievirus B3-infected mice. PLoS Pathog. 2018, 14, e1007235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, A.I.; Grimes, K.A.; Kim, K.; Branche, E.; Bakkenist, C.J.; DePas, W.H.; Shresta, S.; Coyne, C.B. Human FcRn expression and Type I Interferon signaling control Echovirus 11 pathogenesis in mice. PLoS Pathog. 2021, 17, e1009252. [Google Scholar] [CrossRef]
- Ye, N.; Gong, X.; Pang, L.-L.; Gao, W.-J.; Zhang, Y.-T.; Li, X.-L.; Liu, N.; Li, D.-D.; Jin, Y.; Duan, Z.-J. Cytokine responses and correlations thereof with clinical profiles in children with enterovirus 71 infections. BMC Infect. Dis. 2015, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Marco-Puche, G.; Lois, S.; Benítez, J.; Trivino, J.C. RNA-Seq Perspectives to Improve Clinical Diagnosis. Front. Genet. 2019, 10, 1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proceedings of the National Academy of Sciences of the United States of America. Annual subject and author indexes. Proc. Natl. Acad. Sci. USA 1990, 87 (Suppl. 1), 10069–10240. [Google Scholar]
- Cheng, Y.S.; Colonno, R.J.; Yin, F.H. Interferon induction of fibroblast proteins with guanylate binding activity. J. Biol. Chem. 1983, 258, 7746–7750. [Google Scholar] [CrossRef]
- Gupta, S.L.; Rubin, B.Y.; Holmes, S.L. Interferon action: Induction of specific proteins in mouse and human cells by homologous interferons. Proc. Natl. Acad. Sci. USA 1979, 76, 4817–4821. [Google Scholar] [CrossRef] [Green Version]
- Davidson, S.; Maini, M.; Wack, A. Disease-Promoting Effects of Type I Interferons in Viral, Bacterial, and Coinfections. J. Interf. Cytokine Res. 2015, 35, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Guenzi, E.; Töpolt, K.; Cornali, E.; Lubeseder-Martellato, C.; Jörg, A.; Matzen, K.; Zietz, C.; Kremmer, E.; Nappi, F.; Schwemmle, M.; et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001, 20, 5568–5577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenzi, E.; Töpolt, K.; Lubeseder-Martellato, C.; Jörg, A.; Naschberger, E.; Benelli, R.; Albini, A.; Stürzl, M. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003, 22, 3772–3782. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.L.; Carton, J.M.; Lou, J.; Xing, L.; Rubin, B.Y. Interferon-Induced Guanylate Binding Protein-1 (GBP-1) Mediates an Antiviral Effect against Vesicular Stomatitis Virus and Encephalomyocarditis Virus. Virology 1999, 256, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krapp, C.; Hotter, D.; Gawanbacht, A.; McLaren, P.J.; Kluge, S.F.; Stürzel, C.M.; Mack, K.; Reith, E.; Engelhart, S.; Ciuffi, A.; et al. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe 2016, 19, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyrkalska, S.; Candel, S.; Angosto, D.; Gómez-Abellán, V.; Martín-Sánchez, F.; García-Moreno, D.; Zapata-Pérez, R.; Sánchez-Ferrer, A.; Sepulcre, M.P.; Pelegrin, P.; et al. Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat. Commun. 2016, 7, 12077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenoy, A.R.; Wellington, D.A.; Kumar, P.; Kassa, H.; Booth, C.J.; Cresswell, P.; MacMicking, J.D. GBP5 Promotes NLRP3 Inflammasome Assembly and Immunity in Mammals. Science 2012, 336, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Schepart, B.S.; Woodward, J.G.; Palmer, M.J.; Macchi, M.J.; Basta, P.; McLaughlin-Taylor, E.; Frelinger, J.A. Expression in L cells of transfected class I genes from the mouse major histocompatibility complex. Proc. Natl. Acad. Sci. USA 1985, 82, 5505–5509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, T.A. The Role of the Major Histocompatibility Complex in Immunobiology. Proc. R. Soc. Med. 1982, 75, 681. [Google Scholar]
- Rodriguez, F.; Slifka, M.K.; Harkins, S.; Whitton, J.L. Two overlapping subdominant epitopes identified by DNA immunization induce protective CD8(+) T-cell populations with differing cytolytic activities. J. Virol. 2001, 75, 7399–7409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamai, M.; Kobayashi, N.; Shimada, K.; Oka, N.; Takahashi, M.; Tanuma, A.; Kondo, K. Increased interleukin-1β and basic fibroblast growth factor levels in the cerebrospinal fluid during human herpesvirus-6B (HHV-6B) encephalitis. Biochem. Biophys. Res. Commun. 2017, 486, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, T.; Ferreira, H.; Okamura, L.; Giroto, T.; de Oliveira, B.R.S.M.; Fabri, C.; Gameiro, R.; Flores, E. Cellular response markers and cytokine gene expression in the central nervous system of cattle naturally infected with bovine herpesvirus 5. Veter-J. 2016, 218, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Kothur, K.; Wienholt, L.; Mohammad, S.; Tantsis, E.M.; Pillai, S.; Britton, P.N.; Jones, C.A.; Angiti, R.R.; Barnes, E.H.; Schlub, T.; et al. Utility of CSF Cytokine/Chemokines as Markers of Active Intrathecal Inflammation: Comparison of Demyelinating, Anti-NMDAR and Enteroviral Encephalitis. PLoS ONE 2016, 11, e0161656. [Google Scholar] [CrossRef] [PubMed]
- Whitman, L.; Zhou, H.; Perlman, S.; Lane, T.E. IFN-gamma-mediated suppression of coronavirus replication in glial-committed progenitor cells. Virology 2009, 384, 209–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.T.; Stohlman, S.A.; Hinton, D.R. Mouse hepatitis virus is cleared from the central nervous systems of mice lacking perforin-mediated cytolysis. J. Virol. 1997, 71, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, K.; Ulvestad, E.; Cragg, L.; Blain, M.; Antel, J.P. Induction of primary T cell responses by human glial cells. J. Neurosci. Res. 1993, 36, 382–390. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Li, J.; Sun, Q.; Zhang, K.; Xu, W.; Zhang, Y.; Wu, G. Pathological Features of Echovirus-11-Associated Brain Damage in Mice Based on RNA-Seq Analysis. Viruses 2021, 13, 2477. https://doi.org/10.3390/v13122477
Zhang G, Li J, Sun Q, Zhang K, Xu W, Zhang Y, Wu G. Pathological Features of Echovirus-11-Associated Brain Damage in Mice Based on RNA-Seq Analysis. Viruses. 2021; 13(12):2477. https://doi.org/10.3390/v13122477
Chicago/Turabian StyleZhang, Guoyan, Jichen Li, Qiang Sun, Keyi Zhang, Wenbo Xu, Yong Zhang, and Guizhen Wu. 2021. "Pathological Features of Echovirus-11-Associated Brain Damage in Mice Based on RNA-Seq Analysis" Viruses 13, no. 12: 2477. https://doi.org/10.3390/v13122477





