Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primary Cell Isolation
2.2. Flow Cytometric Analysis
2.3. In Vitro Stimulation Assays
2.4. Statistical Analysis
3. Results
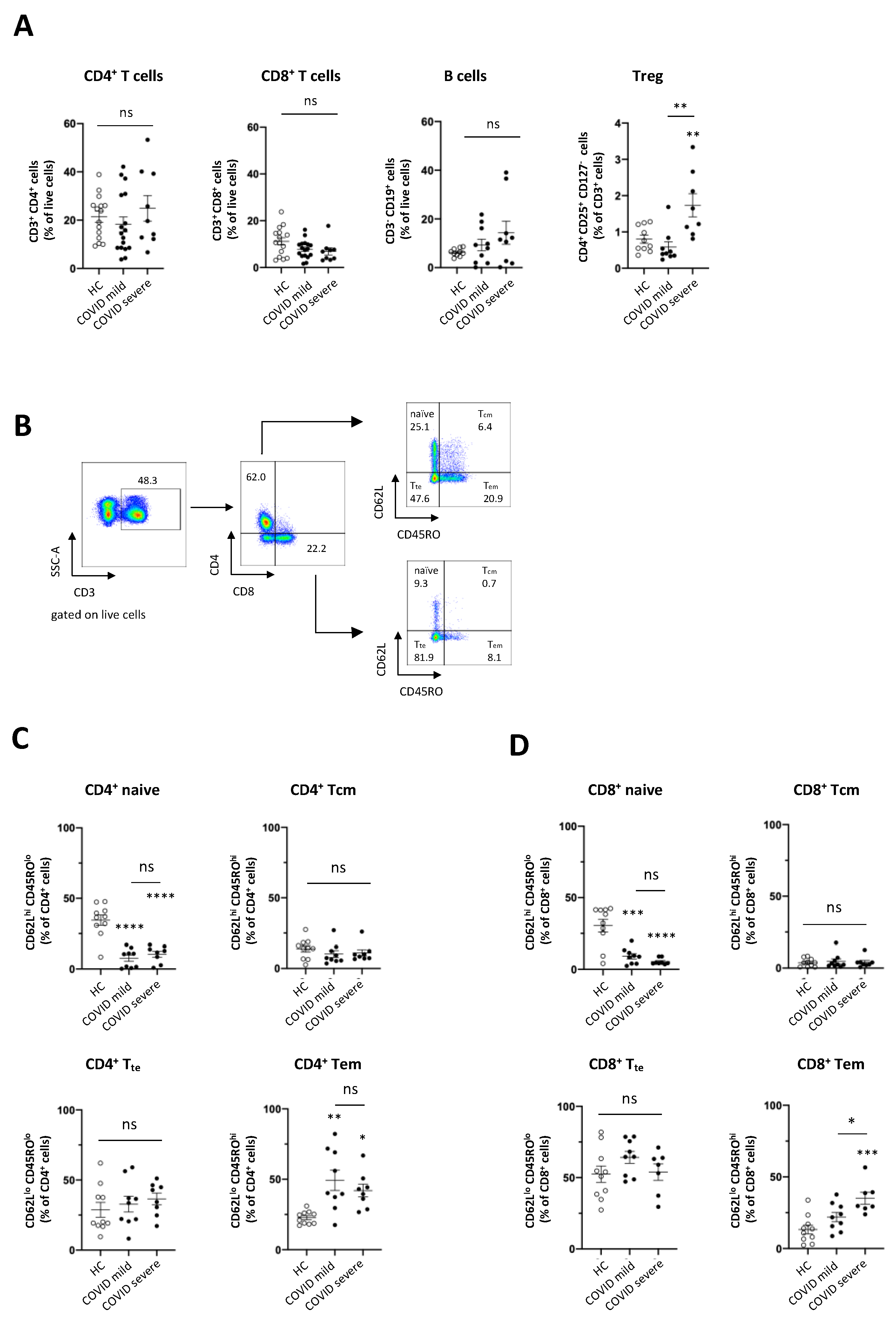
3.1. The Frequency of Adaptive Immune Cell Populations Is Altered in Patients with COVID-19, Irrespective of the Clinical Course of Disease
3.2. MAIT Cells Are Severely Reduced and Phenotypically Altered in Peripheral Blood of Patients with COVID-19
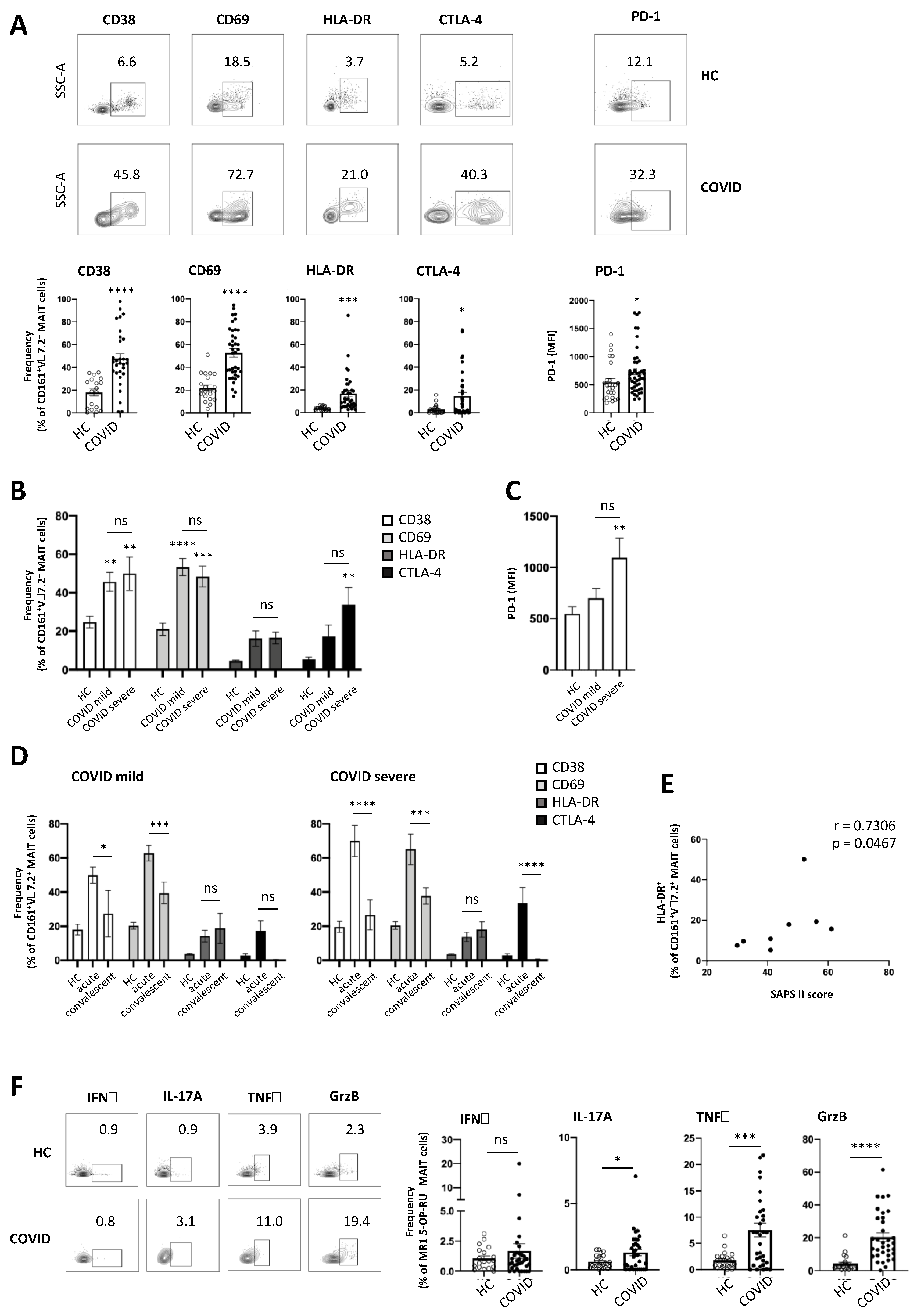
3.3. MAIT Cells Express a Highly Activated Phenotype in COVID-19 Patients
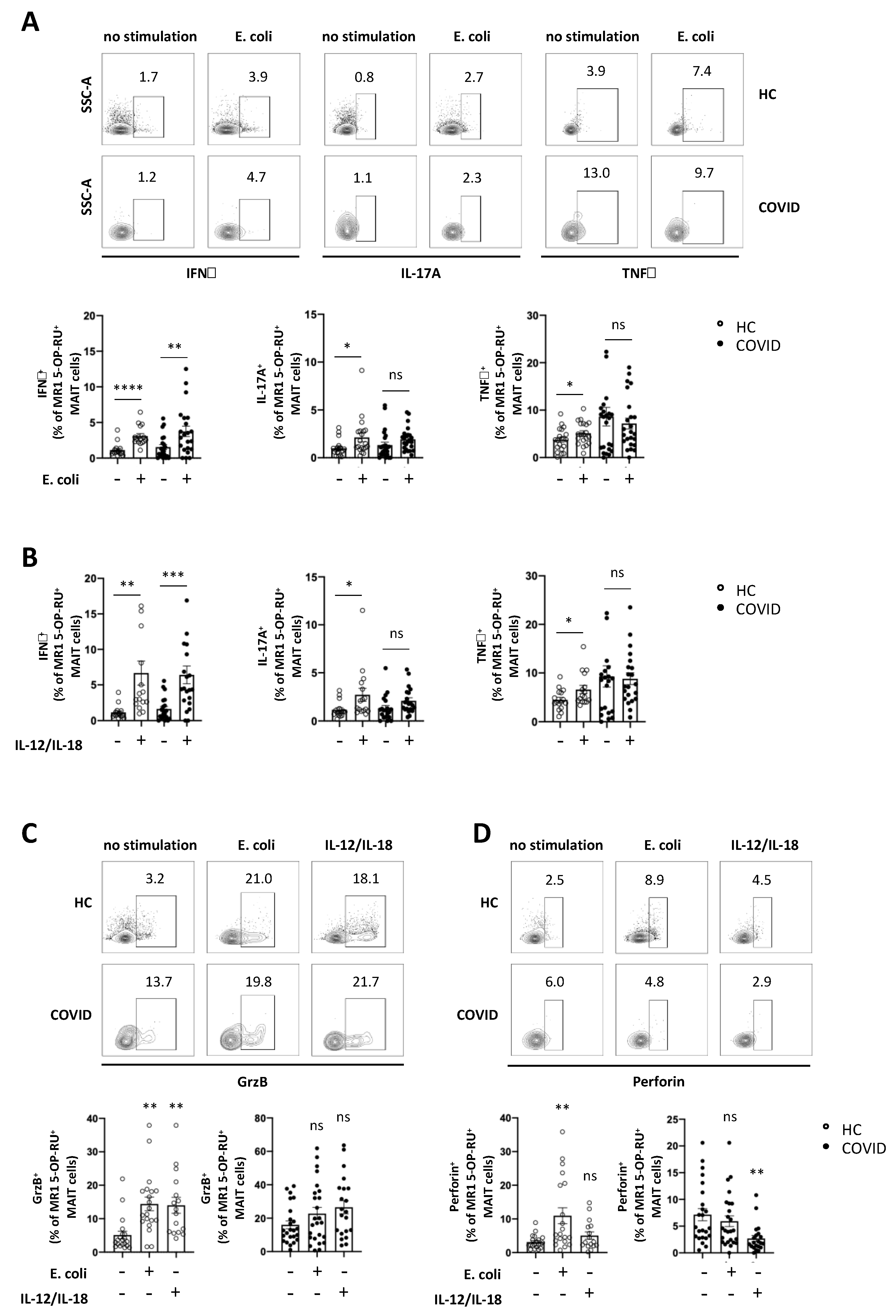
3.4. Expression of Cytokines and Cytolytic Proteins in Response to Specific In Vitro Stimulation is Severely Altered in MAIT Cells from COVID-19 Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARDS | acute respiratory distress syndrome |
| CD | cluster of differentiation |
| COPD | chronic obstructive pulmonary disease |
| COVID-19 | coronavirus disease 2019 |
| CRP | C-reactive protein |
| CTLA-4 | cytotoxic T-lymphocyte-associated protein 4 |
| E. coli | Escherichia coli |
| ECMO | extracorporeal membrane oxygenation |
| HLA-DR | human leukocyte antigen—DR isotype |
| ICU | intensive care unit |
| IFNγ | interferon γ |
| IL | interleukin |
| MAIT cells | mucosal-associated invariant T cells |
| NK cells | natural killer cells |
| PBMCs | peripheral blood mononuclear cells |
| PCR | polymerase chain reaction |
| PD-1 | programmed cell death protein 1 |
| SAPS | Simplified Acute Physiology Score |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SIRS | systemic inflammatory response syndrome |
| SOFA | sepsis-related organ failure assessment |
| TNFα | tumour necrosis factor α |
| WHO | World Health Organisation |
References
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 41, 145–151.
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.G.T.; Fu, H.; et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020, 20, 669–677. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.a.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Siddiqi, H.K.; Mehra, M.R. COVID-19 illness in native and immunosuppressed states: A clinical-therapeutic staging proposal. J. Heart Lung Transpl. 2020, 39, 405–407. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.J.M.; Leong, J.Y.; Lee, J.H.; Albani, S.; Yeo, J.G. Insights into the immuno-pathogenesis of acute respiratory distress syndrome. Ann. Transl. Med. 2019, 7, 504. [Google Scholar] [CrossRef]
- Chowdhury, M.A.; Hossain, N.; Kashem, M.A.; Shahid, M.A.; Alam, A. Immune response in COVID-19: A review. J. Infect. Public Health 2020, 13, 1619–1629. [Google Scholar] [CrossRef]
- Ni, L.; Ye, F.; Cheng, M.-L.; Feng, Y.; Deng, Y.-Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971–977.e3. [Google Scholar] [CrossRef]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2013, 491, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Dusseaux, M.; Martin, E.; Serriari, N.; Peguillet, I.; Premel, V.; Louis, D.; Milder, M.; Le Bourhis, L.; Soudais, C.; Treiner, E.; et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 2011, 117, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Hinks, T.S.C.; Wallington, J.C.; Williams, A.P.; Djukanović, R.; Staples, K.J.; Wilkinson, T.M.A. Steroid-induced Deficiency of Mucosal-associated Invariant T Cells in the Chronic Obstructive Pulmonary Disease Lung. Implications for Nontypeable Haemophilus influenzae Infection. Am. J. Respir. Crit. Care Med. 2016, 194, 1208–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurioka, A.; Ussher, J.E.; Cosgrove, C.; Clough, C.; Fergusson, J.R.; Smith, K.; Kang, Y.H.; Walker, L.J.; Hansen, T.H.; Willberg, C.B.; et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 2014, 8, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Boulouis, C.; Sia, W.R.; Gulam, M.Y.; Teo, J.Q.M.; Png, Y.T.; Phan, T.K.; Mak, J.Y.W.; Fairlie, D.P.; Poon, I.K.H.; Koh, T.H.; et al. Human MAIT cell cytolytic effector proteins synergize to overcome carbapenem resistance in Escherichia coli. PLoS Biol. 2020, 18, e3000644. [Google Scholar] [CrossRef]
- Le Bourhis, L.; Dusseaux, M.; Bohineust, A.; Bessoles, S.; Martin, E.; Premel, V.; Core, M.; Sleurs, D.; Serriari, N.E.; Treiner, E.; et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 2013, 9, e1003681. [Google Scholar] [CrossRef] [Green Version]
- Leeansyah, E.; Svard, J.; Dias, J.; Buggert, M.; Nystrom, J.; Quigley, M.F.; Moll, M.; Sonnerborg, A.; Nowak, P.; Sandberg, J.K. Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection. PLoS Pathog. 2015, 11, e1005072. [Google Scholar] [CrossRef]
- Hartmann, N.; Harriff, M.J.; McMurtrey, C.P.; Hildebrand, W.H.; Lewinsohn, D.M.; Kronenberg, M. Role of MAIT cells in pulmonary bacterial infection. Mol. Immunol. 2018, 101, 155–159. [Google Scholar] [CrossRef]
- Lu, B.; Liu, M.; Wang, J.; Fan, H.; Yang, D.; Zhang, L.; Gu, X.; Nie, J.; Chen, Z.; Corbett, A.J.; et al. IL-17 production by tissue-resident MAIT cells is locally induced in children with pneumonia. Mucosal Immunol. 2020, 13, 824–835. [Google Scholar] [CrossRef] [Green Version]
- Van Wilgenburg, B.; Scherwitzl, I.; Hutchinson, E.C.; Leng, T.; Kurioka, A.; Kulicke, C.; de Lara, C.; Cole, S.; Vasanawathana, S.; Limpitikul, W.; et al. MAIT cells are activated during human viral infections. Nat. Commun. 2016, 7, 11653. [Google Scholar] [CrossRef] [Green Version]
- Ussher, J.E.; Bilton, M.; Attwod, E.; Shadwell, J.; Richardson, R.; de Lara, C.; Mettke, E.; Kurioka, A.; Hansen, T.H.; Klenerman, P.; et al. CD161 ++CD8 +T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 2013, 44, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Van Wilgenburg, B.; Loh, L.; Chen, Z.; Pediongco, T.J.; Wang, H.; Shi, M.; Zhao, Z.; Koutsakos, M.; Nüssing, S.; Sant, S.; et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat. Commun. 2018, 9, 4706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbett, A.J.; Eckle, S.B.G.; Birkinshaw, R.W.; Liu, L.; Patel, O.; Mahony, J.; Chen, Z.; Reantragoon, R.; Meehan, B.; Cao, H.; et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014, 509, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef] [PubMed]
- Jouan, Y.; Guillon, A.; Gonzalez, L.; Perez, Y.; Ehrmann, S.; Ferreira, M.; Daix, T.; Jeannet, R.; Francois, B.; Dequin, P.-F.; et al. Functional alteration of innate T cells in critically ill Covid-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Sutton, C.E.; Mielke, L.A.; Mills, K.H.G. IL-17-producing γδ T cells and innate lymphoid cells. Eur. J. Immunol. 2012, 42, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Belz, G.T.; Denman, R.; Seillet, C.; Jacquelot, N. Tissue-resident lymphocytes: Weaponized sentinels at barrier surfaces. F1000Res 2020, 9. [Google Scholar] [CrossRef]
- Haeryfar, S.M.M. MAIT Cells in COVID-19: Heroes, Villains, or Both? Crit Rev. Immunol 2020, 40, 173–184. [Google Scholar] [CrossRef]
- Walker, L.J.; Kang, Y.-H.; Smith, M.O.; Tharmalingham, H.; Ramamurthy, N.; Fleming, V.M.; Sahgal, N.; Leslie, A.; Oo, Y.; Geremia, A.; et al. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8(+) T cells. Blood 2012, 119, 422–433. [Google Scholar] [CrossRef] [Green Version]
- Lamichhane, R.; Schneider, M.; de la Harpe, S.M.; Harrop, T.W.R.; Hannaway, R.F.; Dearden, P.K.; Kirman, J.R.; Tyndall, J.D.A.; Vernall, A.J.; Ussher, J.E. TCR- or Cytokine-Activated CD8(+) Mucosal-Associated Invariant T Cells Are Rapid Polyfunctional Effectors That Can Coordinate Immune Responses. Cell Rep. 2019, 28, 3061–3076.e5. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J. Clin. Investig. 2020. [Google Scholar] [CrossRef] [Green Version]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Parrot, T.; Gorin, J.B.; Ponzetta, A.; Maleki, K.T.; Kammann, T.; Emgard, J.; Perez-Potti, A.; Sekine, T.; Rivera-Ballesteros, O.; Karolinska, C.-S.G.; et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Flament, H.; Rouland, M.; Beaudoin, L.; Toubal, A.; Bertrand, L.; Lebourgeois, S.; Gouda, Z.; Rousseau, C.; Soulard, P.; Hurtado-Nedelec, M.; et al. Outcome of SARS-CoV-2 infection linked to MAIT cell activation and cytotoxicity: Evidence for an IL-18 dependent mechanism. medRxiv 2020. [Google Scholar] [CrossRef]
- Wang, H.; D’Souza, C.; Lim, X.Y.; Kostenko, L.; Pediongco, T.J.; Eckle, S.B.G.; Meehan, B.S.; Shi, M.; Wang, N.; Li, S.; et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 2018, 9, 3350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zajac, A.J.; Blattman, J.N.; Murali-Krishna, K.; Sourdive, D.J.; Suresh, M.; Altman, J.D.; Ahmed, R. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 1998, 188, 2205–2213. [Google Scholar] [CrossRef]
- Stelekati, E.; Shin, H.; Doering, T.A.; Dolfi, D.V.; Ziegler, C.G.; Beiting, D.P.; Dawson, L.; Liboon, J.; Wolski, D.; Ali, M.-A.A.; et al. Bystander Chronic Infection Negatively Impacts Development of CD8+ T Cell Memory. Immunity 2014, 40, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 131, 492–499. [Google Scholar] [CrossRef]
- Böttcher, K.; Rombouts, K.; Saffioti, F.; Roccarina, D.; Rosselli, M.; Hall, A.; Luong, T.; Tsochatzis, E.A.; Thorburn, D.; Pinzani, M. MAIT cells are chronically activated in patients with autoimmune liver disease and promote pro-fibrogenic hepatic stellate cell activation. Hepatology 2018, 68, 172–186. [Google Scholar] [CrossRef] [Green Version]
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; El Jammal, T.; Walzer, T.; François, B.; Sève, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Meierovics, A.; Yankelevich, W.J.; Cowley, S.C. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, E3119–E3128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meierovics, A.I.; Cowley, S.C. MAIT cells promote inflammatory monocyte differentiation into dendritic cells during pulmonary intracellular infection. J. Exp. Med. 2016, 213, 2793–2809. [Google Scholar] [CrossRef] [Green Version]
- Ginsburg, A.S.; Klugman, K.P. COVID-19 pneumonia and the appropriate use of antibiotics. Lancet Glob. Health 2020, 8, e1453–e1454. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Chien, Y.W.; Klugman, K.P.; Morens, D.M. Bacterial pathogens and death during the 1918 influenza pandemic. N. Engl. J. Med. 2009, 361, 2582–2583. [Google Scholar] [CrossRef] [Green Version]

| COVID-19 Mild | COVID-19 Severe | HC | ||
|---|---|---|---|---|
| Cases (%) | number | 22 (51.2) | 21 (48.8) | 25 |
| Samples (%) | number | 24 (47.1) | 27 (52.9) | n.a. |
| Baseline Characteristics | ||||
| Age (years) | median | 62 | 65 | 28 |
| (range) | (24–86) | (26–84) | (23–41) | |
| Male | number (%) | 12 (54.5) | 16 (76.2) | 13 (52.0) |
| Female | number (%) | 10 (45.5) | 5 (23.8) | 12 (48.0) |
| Positive PCR before sampling | number (%) | 24 (100) | 27 (100) | 0 (0) |
| Symptom onset—sampling (days) | median | 9 | 39.5 | n.a. |
| (range) | (1–56) | (7–74) | ||
| IL-6 (pg/mL) at sampling | median | 12.4 | 55.7 | n.a. |
| (range) | (1.5–101.7) | (5.6–11014) | ||
| CRP (mg/dL) at sampling | median | 2.8 | 5.1 | n.a. |
| (range) | (0.1–10.2) | (0.3–37.3) | ||
| Lymphocyte count total (×109/L) | median | 1.0 | 0.7 | n.a. |
| (range) | (0.1–2.7) | (0.3–2.4) | ||
| Outcome deceased | number (%) | 0 (0) | 9 (42,3) | n.a. |
| Oxygen Supply | n.a. | |||
| Oxygen therapy <15L | number (%) | 6 (27.3) | 2 (9.5) | |
| Noninvasive ventilation | number (%) | 0 (0) | 0 (0) | |
| Invasive ventilation | number (%) | 0 (0) | 19 (90.5) | |
| ECMO | number (%) | 0 (0) | 5 (23.8) | |
| Comorbidities | ||||
| No comorbidity | number (%) | 5 (22.7) | 1 (4.8) | 25 (100.0) |
| Hypertension | number (%) | 9 (40.9) | 14 (66.7) | |
| Diabetes | number (%) | 4 (18.2) | 8 (38.1) | |
| Coronary heart disease | number (%) | 2 (9.1) | 4 (19.0) | |
| COPD/Asthma | number (%) | 4 (18.2) | 1 (4.8) | |
| Chronic kidney disease | number (%) | 0 (0) | 6 (28.6) | |
| Cancer (under treatment) | number (%) | 4 (18.2) | 5 (23.8) | |
| Disease Severity (ICU) | n.a. | |||
| SOFA at admission to ICU | median | n.a. | 11 | |
| (range) | (2–14) | |||
| SAPS II at sampling | median | n.a. | 41 | |
| (range) | (13–61) | |||
| COVID-Specific Treatment | n.a. | |||
| Treatment total | number (%) | 5 (22.7) | 15 (71.4) | |
| - Dexamethason | number (%) | 0 (0) | 8 (38.1) | |
| - Remdesivir | number (%) | 5 (22.7) | 6 (28.6) | |
| - Convalesent plasma | number (%) | 1 (4.5) | 3 (14.3) | |
| - Hydrocortison | number (%) | 0 (0) | 5 (23.8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deschler, S.; Kager, J.; Erber, J.; Fricke, L.; Koyumdzhieva, P.; Georgieva, A.; Lahmer, T.; Wiessner, J.R.; Voit, F.; Schneider, J.; et al. Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients. Viruses 2021, 13, 241. https://doi.org/10.3390/v13020241
Deschler S, Kager J, Erber J, Fricke L, Koyumdzhieva P, Georgieva A, Lahmer T, Wiessner JR, Voit F, Schneider J, et al. Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients. Viruses. 2021; 13(2):241. https://doi.org/10.3390/v13020241
Chicago/Turabian StyleDeschler, Sebastian, Juliane Kager, Johanna Erber, Lisa Fricke, Plamena Koyumdzhieva, Alexandra Georgieva, Tobias Lahmer, Johannes R. Wiessner, Florian Voit, Jochen Schneider, and et al. 2021. "Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients" Viruses 13, no. 2: 241. https://doi.org/10.3390/v13020241
APA StyleDeschler, S., Kager, J., Erber, J., Fricke, L., Koyumdzhieva, P., Georgieva, A., Lahmer, T., Wiessner, J. R., Voit, F., Schneider, J., Horstmann, J., Iakoubov, R., Treiber, M., Winter, C., Ruland, J., Busch, D. H., Knolle, P. A., Protzer, U., Spinner, C. D., ... Böttcher, K. (2021). Mucosal-Associated Invariant T (MAIT) Cells Are Highly Activated and Functionally Impaired in COVID-19 Patients. Viruses, 13(2), 241. https://doi.org/10.3390/v13020241








