Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover
Abstract
1. Background
2. Sialic Acids and Their Biological Significance
3. Sialic Acid Receptors of Emerging and Zoonotic Viruses
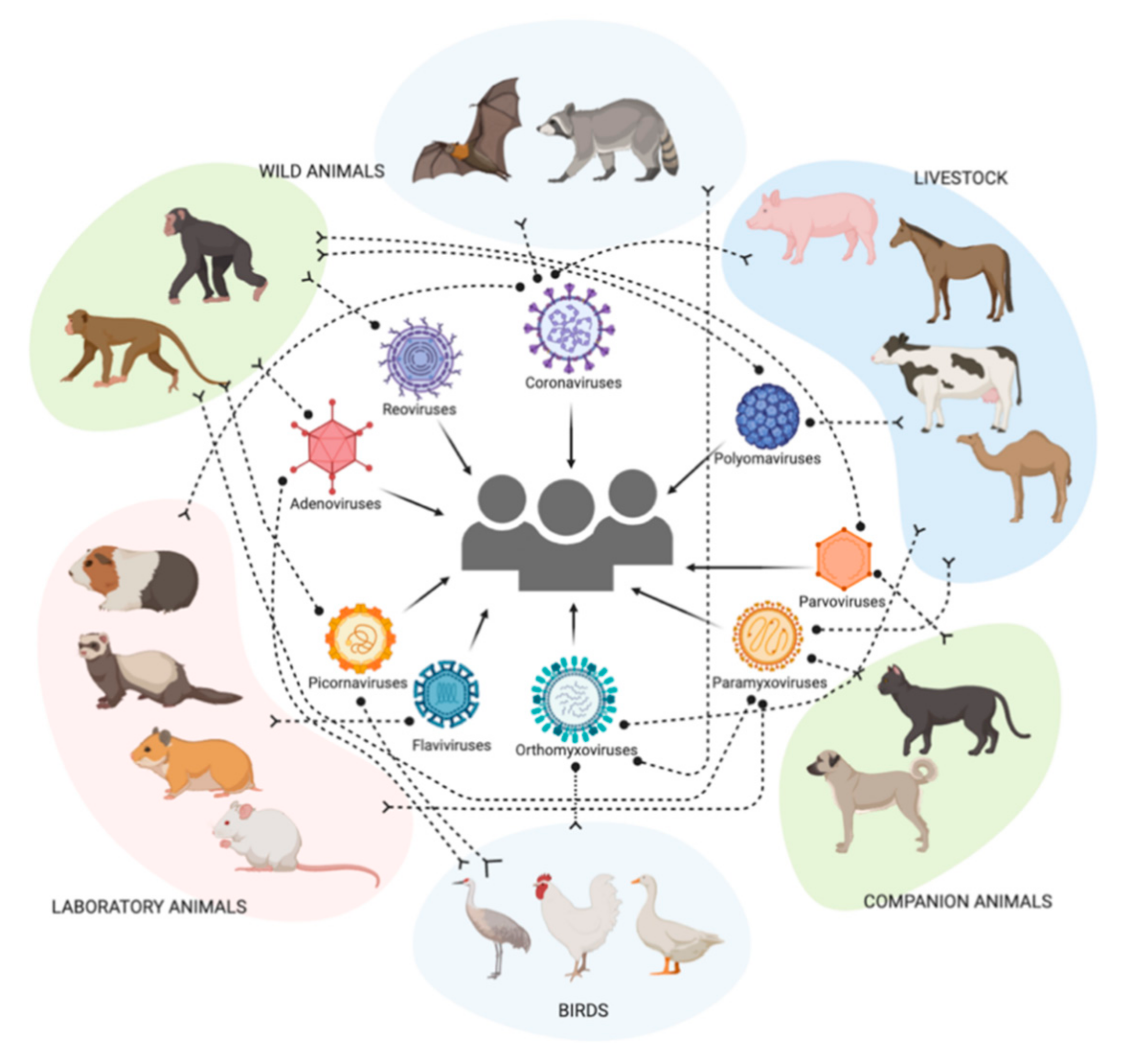
4. Distribution of Sialic Acid Receptors among Multiple Host Species
4.1. Humans
4.2. Non-Human Primates
4.3. Livestock Species and Farm Animals
4.3.1. Swine (Pigs)
4.3.2. Equines (Horses)
4.3.3. Bovines (Cattle)
4.3.4. Camelidae (Camels)
4.4. Companion Animals
4.4.1. Canines (Dogs)
4.4.2. Felines (Cats)
4.5. Wild Animals
4.5.1. Bats (Order: Chiroptera)
4.5.2. Plateau Pika (Ochotona curzoniae)
4.5.3. Raccoon (Procyon lotor)
4.6. Laboratory Animals
4.6.1. Guinea Pig (Cavia porcellus)
4.6.2. Ferrets (Mustela putorius furo)
4.6.3. Hamsters (Mesocricetus auratus)
4.6.4. Mice (Mus musculus)
4.7. Terrestrial Birds
4.7.1. Galliformes
4.7.2. Passeriformes
4.7.3. Columbiformes
4.8. Aquatic Birds
4.8.1. Anseriformes
4.8.2. Charadriiformes
4.8.3. Gruiformes, Pelecaniformes, Gaviiformes, Ciconiiformes
5. SAs as Receptor Determinants for Mammalian Viruses
5.1. RNA Viruses
5.1.1. Orthomyxoviridae
5.1.2. Coronaviridae
5.1.3. Paramyxoviridae
5.1.4. Flaviviridae
5.1.5. Picornaviridae
5.1.6. Reoviridae
5.2. DNA viruses
5.2.1. Adenoviridae
5.2.2. Parvoviridae
5.2.3. Polyomaviridae
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hassell, J.M.; Begon, M.; Ward, M.J.; Fevre, E.M. Urbanization and Disease Emergence: Dynamics at the Wildlife-Livestock-Human Interface. Trends Ecol. Evol. 2017, 32, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.P.; Leibler, J.H.; Price, L.B.; Otte, J.M.; Pfeiffer, D.U.; Tiensin, T.; Silbergeld, E.K. The animal-human interface and infectious disease in industrial food animal production: Rethinking biosecurity and biocontainment. Public Health Rep. 2008, 123, 282–299. [Google Scholar] [CrossRef] [PubMed]
- Maginnis, M.S. Virus-Receptor Interactions: The Key to Cellular Invasion. J. Mol. Biol. 2018, 430, 2590–2611. [Google Scholar] [CrossRef]
- Varki, A.; Gagneux, P. Biological functions of glycans. In Essentials of Glycobiology [Internet], 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017. [Google Scholar]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Varki, N.M.; Varki, A. Diversity in cell surface sialic acid presentations: Implications for biology and disease. Lab. Investig. 2007, 87, 851–857. [Google Scholar] [CrossRef]
- Blix, F.; Gottschalk, A.; Klenk, E. Proposed nomenclature in the field of neuraminic and sialic acids. Nature 1957, 179, 1088. [Google Scholar] [CrossRef]
- Gottschalk, A. The Chemistry and Biology of Sialic Acids and Related Substances; CUP Archive: Cambridge, UK, 1960. [Google Scholar]
- Nadano, D.; Iwasaki, M.; Endo, S.; Kitajima, K.; Inoue, S.; Inoue, Y. A naturally occurring deaminated neuraminic acid, 3-deoxy-D-glycero-D-galacto-nonulosonic acid (KDN). Its unique occurrence at the nonreducing ends of oligosialyl chains in polysialoglycoprotein of rainbow trout eggs. J. Biol. Chem. 1986, 261, 11550–11557. [Google Scholar] [CrossRef]
- Varki, A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 2007, 446, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, F.; Tiralongo, E.; Tiralongo, J. Sialic acid-specific lectins: Occurrence, specificity and function. Cell. Mol. Life Sci. 2006, 63, 1331–1354. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Kelm, S.; Reuter, G.; Roggentin, P.; Shaw, L. Biochemistry and Role of Sialic Acids. In Biology of the Sialic Acids; Rosenberg, A., Ed.; Springer: Boston, MA, USA, 1995; pp. 7–67. [Google Scholar] [CrossRef]
- Vimr, E.R.; Kalivoda, K.A.; Deszo, E.L.; Steenbergen, S.M. Diversity of Microbial Sialic Acid Metabolism. Microbiol. Mol. Biol. Rev. 2004, 68, 132. [Google Scholar] [CrossRef]
- Varki, A.; Gagneux, P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012, 1253, 16–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Brand-Miller, J. The role and potential of sialic acid in human nutrition. Eur. J. Clin. Nutr. 2003, 57, 1351–1369. [Google Scholar] [CrossRef]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology (Read.) 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Wasik, B.R.; Barnard, K.N.; Parrish, C.R. Effects of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol. 2016, 24, 991–1001. [Google Scholar] [CrossRef]
- Haines-Menges, B.L.; Whitaker, W.B.; Lubin, J.B.; Boyd, E.F. Host Sialic Acids: A Delicacy for the Pathogen with Discerning Taste. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Gambaryan, A.; Webster, R.; Matrosovich, M. Differences between influenza virus receptors on target cells of duck and chicken. Arch. Virol. 2002, 147, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- França, M.; Stallknecht, D.E.; Howerth, E.W. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian Pathol. 2013, 42, 60–71. [Google Scholar] [CrossRef]
- Nicholls, J.M.; Chan, R.W.; Russell, R.J.; Air, G.M.; Peiris, J.M. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008, 16, 149–157. [Google Scholar] [CrossRef]
- Geisler, C.; Jarvis, D.L. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 2011, 21, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Akimoto, Y.; Sato, Y.; Kawakami, H.; Hirano, H.; Endo, T. Distribution of Sialoglycoconjugates in the Rat Cerebellum and Its Change with Aging. J. Histochem. Cytochem. 2002, 50, 1179–1186. [Google Scholar] [CrossRef]
- Konami, Y.; Yamamoto, K.; Osawa, T.; Irimura, T. Strong affinity of Maackia amurensis hemagglutinin (MAH) for sialic acid-containing Ser/Thr-linked carbohydrate chains of N-terminal octapeptides from human glycophorin A. FEBS Lett. 1994, 342, 334–338. [Google Scholar] [CrossRef]
- Bateman, A.C.; Karamanska, R.; Busch, M.G.; Dell, A.; Olsen, C.W.; Haslam, S.M. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: The importance of NeuAc{alpha}2-6 glycans. J. Biol. Chem. 2010, 285, 34016–34026. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.W.; Karamanska, R.; Van Poucke, S.; Van Reeth, K.; Chan, I.W.; Chan, M.C.; Dell, A.; Peiris, J.S.; Haslam, S.M.; Guan, Y.; et al. Infection of swine ex vivo tissues with avian viruses including H7N9 and correlation with glycomic analysis. Influenza Other Respir. Viruses 2013, 7, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Nishikaze, T. Sialic acid derivatization for glycan analysis by mass spectrometry. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Nelli, R.; White, G.A.; Bain, M.; Chang, K.C.; Dunham, S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. J. Mol. Genet. Med. 2009, 3, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.; Karamanska, R.; Chan, R.W.; Chan, M.C.; Jia, N.; Air, G.; Hopton, C.; Wong, M.P.; Dell, A.; Malik Peiris, J.S.; et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013, 9, e1003223. [Google Scholar] [CrossRef] [PubMed]
- Byrd-Leotis, L.; Liu, R.; Bradley, K.C.; Lasanajak, Y.; Cummings, S.F.; Song, X.; Heimburg-Molinaro, J.; Galloway, S.E.; Culhane, M.R.; Smith, D.F.; et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc. Natl. Acad. Sci. USA 2014, 111, E2241–E2250. [Google Scholar] [CrossRef] [PubMed]
- Siegers, J.Y.; van de Bildt, M.W.G.; Lin, Z.; Leijten, L.M.; Lavrijssen, R.A.M.; Bestebroer, T.; Spronken, M.I.J.; De Zeeuw, C.I.; Gao, Z.; Schrauwen, E.J.A.; et al. Viral Factors Important for Efficient Replication of Influenza A Viruses in Cells of the Central Nervous System. J. Virol. 2019, 93, e02273-18. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, M.; Fouchier, R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014, 33, 823–841. [Google Scholar] [CrossRef]
- Löfling, J.; Lyi, S.M.; Parrish, C.R.; Varki, A. Canine and feline parvoviruses preferentially recognize the non-human cell surface sialic acid N-glycolylneuraminic acid. Virology 2013, 440, 89–96. [Google Scholar] [CrossRef]
- Seiradake, E.; Henaff, D.; Wodrich, H.; Billet, O.; Perreau, M.; Hippert, C.; Mennechet, F.; Schoehn, G.; Lortat-Jacob, H.; Dreja, H.; et al. The Cell Adhesion Molecule “CAR” and Sialic Acid on Human Erythrocytes Influence Adenovirus in Vivo Biodistribution. PLoS Pathog. 2009, 5, e1000277. [Google Scholar] [CrossRef]
- Yao, L.; Korteweg, C.; Hsueh, W.; Gu, J. Avian influenza receptor expression in H5N1-infected and noninfected human tissues. FASEB J. 2008, 22, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Altheide, T.K.; Varki, A. A uniquely human consequence of domain-specific functional adaptation in a sialic acid-binding receptor. Glycobiology 2004, 14, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Ebina, M.; Yamada, S.; Ono, M.; Kasai, N.; Kawaoka, Y. Avian flu: Influenza virus receptors in the human airway. Nature 2006, 440, 435–436. [Google Scholar] [CrossRef]
- Sata, T.; Roth, J.; Zuber, C.; Stamm, B.; Heitz, P.U. Expression of alpha 2,6-linked sialic acid residues in neoplastic but not in normal human colonic mucosa. A lectin-gold cytochemical study with Sambucus nigra and Maackia amurensis lectins. Am. J. Pathol. 1991, 139, 1435–1448. [Google Scholar]
- Bertram, S.; Heurich, A.; Lavender, H.; Gierer, S.; Danisch, S.; Perin, P.; Lucas, J.M.; Nelson, P.S.; Pöhlmann, S.; Soilleux, E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE 2012, 7, e35876. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.-D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Suguitan, A.; Orandle, M.; Paskel, M.; Boonnak, K.; Gardner, D.J.; Feldmann, F.; Feldmann, H.; Marino, M.; Jin, H.; et al. African Green Monkeys Recapitulate the Clinical Experience with Replication of Live Attenuated Pandemic Influenza Virus Vaccine Candidates. J. Virol. 2014, 88, 8139–8152. [Google Scholar] [CrossRef]
- Nelli, R.K.; Kuchipudi, S.V.; White, G.A.; Perez, B.B.; Dunham, S.P.; Chang, K.C. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 2010, 6. [Google Scholar] [CrossRef]
- Eriksson, P.; Lindskog, C.; Engholm, E.; Blixt, O.; Waldenström, J.; Munster, V.; Lundkvist, Å; Olsen, B.; Jourdain, E.; Ellström, P. Characterization of avian influenza virus attachment patterns to human and pig tissues. Sci. Rep. 2018, 8, 12215. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.; Thomson, P.C.; Wynn, P.C.; Wang, B. The non-human glycan, N-glycolylneuraminic acid (Neu5Gc), is not expressed in all organs and skeletal muscles of nine animal species. Food Chem. 2021, 343, 128439. [Google Scholar] [CrossRef] [PubMed]
- Muranaka, M.; Yamanaka, T.; Katayama, Y.; Hidari, K.; Kanazawa, H.; Suzuki, T.; Oku, K.; Oyamada, T. Distribution of Influenza Virus Sialoreceptors on Upper and Lower Respiratory Tract in Horses and Dogs. J. Vet. Med. Sci. 2011, 73, 125–127. [Google Scholar] [CrossRef][Green Version]
- Thontiravong, A.; Rung-ruangkijkrai, T.; Kitikoon, P.; Oraveerakul, K.; Poovorawan, Y. Influenza A Virus Receptor Identification in the Respiratory Tract of Quail, Pig, Cow and Swamp Buffalo. Thai J. Vet. Med. 2011, 41, 371–376. [Google Scholar]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog. 2016, 12, e1005411. [Google Scholar] [CrossRef]
- Wasik, B.R.; Barnard, K.N.; Ossiboff, R.J.; Khedri, Z.; Feng, K.H.; Yu, H.; Chen, X.; Perez, D.R.; Varki, A.; Parrish, C.R. Distribution of O-Acetylated Sialic Acids among Target Host Tissues for Influenza Virus. mSphere 2017, 2, e00379-16. [Google Scholar] [CrossRef]
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; van Dieren, B.; Lang, Y.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [PubMed]
- Thongratsakul, S.; Suzuki, Y.; Hiramatsu, H.; Sakpuaram, T.; Sirinarumitr, T.; Poolkhet, C.; Moonjit, P.; Yodsheewan, R.; Songserm, T. Avian and human influenza A virus receptors in trachea and lung of animals. Asian Pac. J. Allergy Immunol. 2010, 28, 294–301. [Google Scholar] [PubMed]
- Ning, Z.-Y.; Wu, X.-T.; Cheng, Y.-F.; Qi, W.-B.; An, Y.-F.; Wang, H.; Zhang, G.-H.; Li, S.-J. Tissue distribution of sialic acid-linked influenza virus receptors in beagle dogs. J. Vet. Sci. 2012, 13, 219–222. [Google Scholar] [CrossRef]
- Jayaraman, A.; Chandrasekaran, A.; Viswanathan, K.; Raman, R.; Fox, J.G.; Sasisekharan, R. Decoding the Distribution of Glycan Receptors for Human-Adapted Influenza A Viruses in Ferret Respiratory Tract. PLoS ONE 2012, 7, e27517. [Google Scholar] [CrossRef] [PubMed]
- Chothe, S.K.; Bhushan, G.; Nissly, R.H.; Yeh, Y.-T.; Brown, J.; Turner, G.; Fisher, J.; Sewall, B.J.; Reeder, D.M.; Terrones, M.; et al. Avian and human influenza virus compatible sialic acid receptors in little brown bats. Sci. Rep. 2017, 7, 660. [Google Scholar] [CrossRef] [PubMed]
- Slater, T.; Eckerle, I.; Chang, K.-C. Bat lung epithelial cells show greater host species-specific innate resistance than MDCK cells to human and avian influenza viruses. Virol. J. 2018, 15, 68. [Google Scholar] [CrossRef]
- Ibricevic, A.; Pekosz, A.; Walter, M.J.; Newby, C.; Battaile, J.T.; Brown, E.G.; Holtzman, M.J.; Brody, S.L. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006, 80, 7469–7480. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, H.; Huang, C.; Sun, H.; Li, L.; Su, J.; Ma, J.; Liu, D.; Wang, H.; Liu, W.; et al. Distribution of sialic acid receptors and experimental infections with different subtypes of influenza A viruses in Qinghai-Tibet plateau wild pika. Virol. J. 2015, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.S.; Bentler, K.T.; Landolt, G.; Elmore, S.A.; Minnis, R.B.; Campbell, T.A.; Barras, S.C.; Root, J.J.; Pilon, J.; Pabilonia, K. Influenza infection in wild raccoons. Emerg. Infect. Dis. 2008, 14, 1842. [Google Scholar] [CrossRef]
- Sun, Y.; Bi, Y.; Pu, J.; Hu, Y.; Wang, J.; Gao, H.; Liu, L.; Xu, Q.; Tan, Y.; Liu, M.; et al. Guinea Pig Model for Evaluating the Potential Public Health Risk of Swine and Avian Influenza Viruses. PLoS ONE 2010, 5, e15537. [Google Scholar] [CrossRef]
- Leigh, M.W.; Connor, R.J.; Kelm, S.; Baum, L.G.; Paulson, J.C. Receptor specificity of influenza virus influences severity of illness in ferrets. Vaccine 1995, 13, 1468–1473. [Google Scholar] [CrossRef]
- Kirkeby, S.; Martel, C.J.; Aasted, B. Infection with human H1N1 influenza virus affects the expression of sialic acids of metaplastic mucous cells in the ferret airways. Virus Res. 2009, 144, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Newby, C.M.; Rowe, R.K.; Pekosz, A. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology 2006, 354, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki-Horimoto, K.; Nakajima, N.; Ichiko, Y.; Sakai-Tagawa, Y.; Noda, T.; Hasegawa, H.; Kawaoka, Y. Syrian Hamster as an Animal Model for the Study of Human Influenza Virus Infection. J. Virol. 2018, 92, e01693-17. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.Y.; Luo, M.Y.; Qi, W.B.; Yu, B.; Jiao, P.R.; Liao, M. Detection of expression of influenza virus receptors in tissues of BALB/c mice by histochemistry. Vet. Res. Commun. 2009, 33, 895–903. [Google Scholar] [CrossRef]
- Kimble, B.; Nieto, G.R.; Perez, D.R. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J. 2010, 7, 365. [Google Scholar] [CrossRef]
- Eriksson, P.; Lindskog, C.; Lorente-Leal, V.; Waldenström, J.; González-Acuna, D.; Järhult, J.D.; Lundkvist, Å.; Olsen, B.; Jourdain, E.; Ellström, P. Attachment Patterns of Human and Avian Influenza Viruses to Trachea and Colon of 26 Bird Species—Support for the Community Concept. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Costa, T.; Chaves, A.J.; Valle, R.; Darji, A.; van Riel, D.; Kuiken, T.; Majó, N.; Ramis, A. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet. Res. 2012, 43, 28. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.M.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D.M.E. Characterization of a Novel Influenza A Virus Hemagglutinin Subtype (H16) Obtained from Black-Headed Gulls. J. Virol. 2005, 79, 2814–2822. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.J.; Kawaoka, Y.; Webster, R.G.; Paulson, J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994, 205, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.N.; D’Souza, B.L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 1989, 173, 317–322. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Baum, L.G.; Paulson, J.C. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem. Suppl. 1990, 40, 35–38. [Google Scholar]
- Liu, J.; Stevens, D.J.; Haire, L.F.; Walker, P.A.; Coombs, P.J.; Russell, R.J.; Gamblin, S.J.J. Structures of receptor complexes formed byhemagglutinins from the Asian Influenza pandemic of1957. Proc. Natl. Acad. Sci. USA 2009, 106, 17175–17180. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; McBride, R.; Paulson, J.C.; Basler, C.F.; Wilson, I.A. Structure, receptorbinding, and antigenicity of influenza virus hemagglutinins from the1957H2N2pandemic. J. Virol. 2010, 84, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Gamblin, S.J.; Haire, L.F.; Russell, R.J.; Stevens, D.J.; Xiao, B.; Ha, Y.; Vasisht, N.; Steinhauer, D.A.; Daniels, R.S.; Elliot, A.; et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 2004, 303, 1838–1842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shi, Y.; Qi, J.; Gao, F.; Li, Q.; Fan, Z.; Yan, J.; Gao, G.F. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J. Virol. 2013, 87, 5949–5958. [Google Scholar] [CrossRef] [PubMed]
- Belser, J.A.; Gustin, K.M.; Pearce, M.B.; Maines, T.R.; Zeng, H.; Pappas, C.; Sun, X.; Carney, P.J.; Villanueva, J.M.; Stevens, J.; et al. Pathogenesisand transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 2013, 501, 556–559. [Google Scholar] [CrossRef]
- Kreijtz, J.H.; Kroeze, E.J.; Stittelaar, K.J.; de Waal, L.; van Amerongen, G.; van Trierum, S.; van Run, P.; Bestebroer, T.; Kuiken, T.; Fouchier, R.A.; et al. Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: A model to study intervention strategies. Vaccine 2013, 31, 4995–4999. [Google Scholar] [CrossRef]
- Watanabe, T.; Kiso, M.; Fukuyama, S.; Nakajima, N.; Imai, M.; Yamada, S.; Murakami, S.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Sakoda, Y.; et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013, 501, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Poon, L.L.M.; Lau, A.S.; Luk, W.; Lau, Y.L.; Shortridge, K.F.; Gordon, S.; Guan, Y.; Peiris, J.S.M. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: A mechanism for the unusual severity of human disease? Lancet 2002, 360, 1831–1837. [Google Scholar] [CrossRef]
- Chan, M.C.W.; Cheung, C.Y.; Chui, W.H.; Tsao, S.W.; Nicholls, J.M.; Chan, Y.O.; Chan, R.W.Y.; Long, H.T.; Poon, L.L.M.; Guan, Y.; et al. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 2005, 6, 135. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Tellabati, M.; Sebastian, S.; Londt, B.Z.; Jansen, C.; Vervelde, L.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; Chang, K.C. Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and pro-inflammatory responses. Vet. Res. 2014, 45, 118. [Google Scholar] [CrossRef]
- De Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.; Chau, T.N.; Hoang, D.M.; Chau, N.V.; Khanh, T.H.; Dong, V.C.; et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.; Bernal-Rubio, D.; Durham, N.; Belicha-Villanueva, A.; Lowen, A.C.; Steel, J.; Fernandez-Sesma, A. Effects of Receptor Binding Specificity of Avian Influenza Virus on the Human Innate Immune Response. J. Virol. 2011, 85, 4421–4431. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Watanabe, T.; Kiso, M.; Nakajima, N.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Hatta, M.; Yamada, S.; Ito, M.; Sakai-Tagawa, Y.; et al. A Highly Pathogenic Avian H7N9 Influenza Virus Isolated from A Human Is Lethal in Some Ferrets Infected via Respiratory Droplets. Cell Host Microbe 2017, 22, 615–626.e618. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Schrauwen, E.J.A.; Graaf, M.D.; Bestebroer, T.M.; Spronken, M.; Boheemen, S.V.; Meulder, D.D.; Lexmond, P.; Linster, M.; Herfst, S.; et al. Limited airborne transmission of influenza A/H7N9 virus between ferrets. Nature 2013, 501, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, D.; Kelvin, D.J.; Li, L.; Zheng, Z.; Yoon, S.W.; Wong, S.S.; Farooqui, A.; Wang, J.; Banner, D.; et al. Infectivity, transmission, and pathology of human-isolated H7N9influenza virus inferrets and pigs. Science 2013, 341, 183–186. [Google Scholar] [CrossRef]
- Ni, F.; Nnadi Mbawuike, I.; Kondrashkina, E.; Wang, Q. The roles of hemagglutinin Phe-95 in receptor binding and pathogenicity of influenza B virus. Virology 2014, 450–451, 71–83. [Google Scholar] [CrossRef]
- Horimoto, T.; Gen, F.; Murakami, S.; Iwatsuki-Horimoto, K.; Kato, K.; Akashi, H.; Hisasue, M.; Sakaguchi, M.; Kawaoka, Y.; Maeda, K. Serological evidence of infection of dogs with human influenza viruses in Japan: TABLE 1. Vet. Rec. 2014, 174, 96. [Google Scholar] [CrossRef]
- Manuguerra, J.C.; Hannoun, C.; Simón, F.; Villar, E.; Cabezas, J.A. Natural infection of dogs by influenza C virus: A serological survey in Spain. New Microbiol. 1993, 16, 367–371. [Google Scholar]
- Ohwada, K.; Kitame, F.; Sugawara, K.; Nishimura, H.; Homma, M.; Nakamura, K. Distribution of the Antibody to Influenza C Virus in Dogs and Pigs in Yamagata Prefecture, Japan. Microbiol. Immunol. 1987, 31, 1173–1180. [Google Scholar] [CrossRef]
- Yamaoka, M.; Hotta, H.; Itoh, M.; Homma, M. Prevalence of antibody to influenza C virus among pigs in Hyogo Prefecture, Japan. J. Gen. Virol. 1991, 72, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Porter, E.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Influenza C Virus in Cattle with Respiratory Disease, United States, 2016–2018. Emerg. Infect. Dis. 2018, 24, 1926–1929. [Google Scholar] [CrossRef]
- Nissly, R.H.; Zaman, N.; Ibrahim, P.A.S.; McDaniel, K.; Lim, L.; Kiser, J.N.; Bird, I.; Chothe, S.K.; Bhushan, G.L.; Vandegrift, K.; et al. Influenza C and D viral load in cattle correlates with bovine respiratory disease (BRD): Emerging role of orthomyxoviruses in the pathogenesis of BRD. Virology 2020, 551, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Hellebø, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious Salmon Anemia Virus Specifically Binds to and Hydrolyzes 4-O-Acetylated Sialic Acids. J. Virol. 2004, 78, 3055. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Zhang, S.; Cui, Y.; Shi, Y.; Gao, G.F.; Qi, J. Structures of human-infecting Thogotovirus fusogens support a common ancestor with insect baculovirus. Proc. Natl. Acad. Sci. USA 2017, 114, E8905. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Montero, J.V.; Soriano, V.; Barreiro, P.; de Mendoza, C.; Artacho, M.A. Coronavirus and other airborne agents with pandemic potential. Curr. Opin. Environ. Sci. Health 2020, 17, 41–48. [Google Scholar] [CrossRef]
- Wielgat, P.; Rogowski, K.; Godlewska, K.; Car, H. Coronaviruses: Is Sialic Acid a Gate to the Eye of Cytokine Storm? From the Entry to the Effects. Cells 2020, 9, 1963. [Google Scholar] [CrossRef]
- Morniroli, D.; Giannì, M.L.; Consales, A.; Pietrasanta, C.; Mosca, F. Human Sialome and Coronavirus Disease-2019 (COVID-19) Pandemic: An Understated Correlation? Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.J.; Bosch, B.J.; Rey, F.A.; de Groot, R.J.; et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Archer-Hartmann, S.; Supekar, N.T.; Gleinich, A.S.; Heiss, C.; Azadi, P. Comprehensive characterization of N- and O-glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic Acid Receptors of Viruses. In SialoGlyco Chemistry and Biology II; Gerardy-Schahn, R., Delannoy, P., von Itzstein, M., Eds.; Springer: Berlin, Germany, 2013; pp. 1–28. [Google Scholar]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.J.; Bosch, B.J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Krempl, C.; Schultze, B.; Laude, H.; Herrler, G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997, 71, 3285–3287. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Yang, Z.; Song, H.; Wang, K.; Yang, Y.; Xie, L.; Huang, S.; Liu, J.; Ran, L.; Song, Z. Three Main Inducers of Alphacoronavirus Infection of Enterocytes: Sialic Acid, Proteases, and Low pH. Intervirology 2018, 61, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Hulswit, R.J.; de Haan, C.A.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Jaimes, J.A.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Qing, E.; Hantak, M.; Perlman, S.; Gallagher, T. Distinct Roles for Sialoside and Protein Receptors in Coronavirus Infection. mBio 2020, 11. [Google Scholar] [CrossRef]
- Kruse, H.; Kirkemo, A.M.; Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 2004, 10, 2067–2072. [Google Scholar] [CrossRef] [PubMed]
- Sheshberadaran, H.; Norrby, E.; McCullough, K.C.; Carpenter, W.C.; Orvell, C. The antigenic relationship between measles, canine distemper and rinderpest viruses studied with monoclonal antibodies. J. Gen. Virol. 1986, 67 Pt 7, 1381–1392. [Google Scholar] [CrossRef]
- Marsh, G.A.; Wang, L.-F. Hendra and Nipah viruses: Why are they so deadly? Curr. Opin. Virol. 2012, 2, 242–247. [Google Scholar] [CrossRef]
- Suzuki, T.; Portner, A.; Scroggs, R.A.; Uchikawa, M.; Koyama, N.; Matsuo, K.; Suzuki, Y.; Takimoto, T. Receptor Specificities of Human Respiroviruses. J. Virol. 2001, 75, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Villar, E.; Barroso, I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: A minireview. Glycoconj. J. 2006, 23, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Kubota, M.; Takeuchi, K.; Watanabe, S.; Ohno, S.; Matsuoka, R.; Kohda, D.; Nakakita, S.-i.; Hiramatsu, H.; Suzuki, Y.; Nakayama, T.; et al. Trisaccharide containing α2,3-linked sialic acid is a receptor for mumps virus. Proc. Natl. Acad. Sci. USA 2016, 113, 11579–11584. [Google Scholar] [CrossRef] [PubMed]
- Bieringer, M.; Han, J.W.; Kendl, S.; Khosravi, M.; Plattet, P.; Schneider-Schaulies, J. Experimental Adaptation of Wild-Type Canine Distemper Virus (CDV) to the Human Entry Receptor CD150. PLoS ONE 2013, 8, e57488. [Google Scholar] [CrossRef]
- Zeltina, A.; Bowden, T.A.; Lee, B. Emerging Paramyxoviruses: Receptor Tropism and Zoonotic Potential. PLoS Pathog. 2016, 12, e1005390. [Google Scholar] [CrossRef]
- Laureti, M.; Narayanan, D.; Rodriguez-Andres, J.; Fazakerley, J.K.; Kedzierski, L. Flavivirus Receptors: Diversity, Identity, and Cell Entry. Front. Immunol. 2018, 9, 2180. [Google Scholar] [CrossRef]
- Tan, C.W.; Huan Hor, C.H.; Kwek, S.S.; Tee, H.K.; Sam, I.C.; Goh, E.L.K.; Ooi, E.E.; Chan, Y.F.; Wang, L.F. Cell surface α2,3-linked sialic acid facilitates Zika virus internalization. Emerg. Microbes Infect. 2019, 8, 426–437. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus entry receptors: An update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef]
- Cime-Castillo, J.; Delannoy, P.; Mendoza-Hernández, G.; Monroy-Martínez, V.; Harduin-Lepers, A.; Lanz-Mendoza, H.; Hernández-Hernández Fde, L.; Zenteno, E.; Cabello-Gutiérrez, C.; Ruiz-Ordaz, B.H. Sialic acid expression in the mosquito Aedes aegypti and its possible role in dengue virus-vector interactions. Biomed. Res. Int. 2015, 2015, 504187. [Google Scholar] [CrossRef]
- Espinosa, D.A.; Beatty, P.R.; Puerta-Guardo, H.; Islam, M.N.; Belisle, J.T.; Perera, R.; Harris, E. Increased serum sialic acid is associated with morbidity and mortality in a murine model of dengue disease. J. Gen. Virol. 2019, 100, 1515–1522. [Google Scholar] [CrossRef]
- Rocklov, J.; Dubrow, R. Climate change: An enduring challenge for vector-borne disease prevention and control. Nat. Immunol. 2020, 21, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T.; Okamoto, M.; Nakakita, S.I.; Suzuki, A.; Saito, M.; Tamaki, R.; Lupisan, S.; Roy, C.N.; Hiramatsu, H.; Sugawara, K.E.; et al. Antigenic and Receptor Binding Properties of Enterovirus 68. J. Virol. 2014, 88, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.C.; Jamshidi, F.; Johansson, S.M.C.; Oberste, M.S.; Arnberg, N. Sialic Acid Is a Cellular Receptor for Coxsackievirus A24 Variant, an Emerging Virus with Pandemic Potential. J. Virol. 2008, 82, 3061–3068. [Google Scholar] [CrossRef] [PubMed]
- Nokhbeh, M.R.; Hazra, S.; Alexander, D.A.; Khan, A.; McAllister, M.; Suuronen, E.J.; Griffith, M.; Dimock, K. Enterovirus 70 Binds to Different Glycoconjugates Containing α2,3-Linked Sialic Acid on Different Cell Lines. J. Virol. 2005, 79, 7087–7094. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.W.; Strauss, G.H.; Langford, M.P. Acute hemorrhagic conjunctivitis. Am. Fam. Physician 1992, 45, 173–178. [Google Scholar]
- Blaum, B.S.; Stehle, T. Sialic Acids in Nonenveloped Virus Infections. Adv. Carbohydr. Chem. Biochem. 2019. [Google Scholar] [CrossRef]
- Mistry, N.; Inoue, H.; Jamshidi, F.; Storm, R.J.; Oberste, M.S.; Arnberg, N. Coxsackievirus A24 Variant Uses Sialic Acid-Containing O-Linked Glycoconjugates as Cellular Receptors on Human Ocular Cells. J. Virol. 2011, 85, 11283–11290. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Feeroz, M.M.; Maher, K.; Nix, W.A.; Engel, G.A.; Begum, S.; Hasan, K.M.; Oh, G.; Pallansch, M.A.; Jones-Engel, L. Naturally Acquired Picornavirus Infections in Primates at the Dhaka Zoo. J. Virol. 2013, 87, 572. [Google Scholar] [CrossRef][Green Version]
- Du, J.; Lu, L.; Liu, F.; Su, H.; Dong, J.; Sun, L.; Zhu, Y.; Ren, X.; Yang, F.; Guo, F.; et al. Distribution and characteristics of rodent picornaviruses in China. Sci. Rep. 2016, 6, 34381. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Cao, S.; Holtz, L.R.; Antonio, M.; Stine, O.C.; Wang, D. Discovery of rosavirus 2, a novel variant of a rodent-associated picornavirus, in children from the Gambia. Virology 2014, 454–455, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Li, K.S.M.; Fan, R.Y.Y.; Bai, R.; Huang, Y.; Chan, K.H.; Yuen, K.Y. Chickens host diverse picornaviruses originated from potential interspecies transmission with recombination. J. Gen. Virol. 2014, 95, 1929–1944. [Google Scholar] [CrossRef]
- Campbell, J.A.; Schelling, P.; Wetzel, J.D.; Johnson, E.M.; Forrest, J.C.; Wilson, G.A.R.; Aurrand-Lions, M.; Imhof, B.A.; Stehle, T.; Dermody, T.S. Junctional Adhesion Molecule A Serves as a Receptor for Prototype and Field-Isolate Strains of Mammalian Reovirus. J. Virol. 2005, 79, 7967–7978. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, E.; Guglielmi, K.M.; Strauss, H.M.; Dermody, T.S.; Stehle, T. Structure of Reovirus σ1 in Complex with Its Receptor Junctional Adhesion Molecule-A. PLoS Pathog. 2008, 4, e1000235. [Google Scholar] [CrossRef]
- Forrest, J.C.; Dermody, T.S. Reovirus Receptors and Pathogenesis. J. Virol. 2003, 77, 9109. [Google Scholar] [CrossRef]
- Chua, K.B.; Voon, K.; Yu, M.; Keniscope, C.; Abdul Rasid, K.; Wang, L.-F. Investigation of a Potential Zoonotic Transmission of Orthoreovirus Associated with Acute Influenza-Like Illness in an Adult Patient. PLoS ONE 2011, 6, e25434. [Google Scholar] [CrossRef] [PubMed]
- Chandra, N.; Frängsmyr, L.; Imhof, S.; Caraballo, R.; Elofsson, M.; Arnberg, N. Sialic Acid-Containing Glycans as Cellular Receptors for Ocular Human Adenoviruses: Implications for Tropism and Treatment. Viruses 2019, 11, 395. [Google Scholar] [CrossRef]
- Borkenhagen, L.K.; Fieldhouse, J.K.; Seto, D.; Gray, G.C. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg. Microbes Infect. 2019, 8, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, S.; Seto, J.; Liu, E.B.; Ismail, A.M.; Madupu, R.; Heim, A.; Jones, M.S.; Dyer, D.W.; Chodosh, J.; Seto, D. A Zoonotic Adenoviral Human Pathogen Emerged through Genomic Recombination among Human and Nonhuman Simian Hosts. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.W.; Tse, H.; Chan, W.-M.; Choi, G.K.Y.; Zhang, A.J.X.; Sridhar, S.; Wong, S.C.Y.; Chan, J.F.W.; Chan, A.S.F.; Woo, P.C.Y.; et al. A Novel Psittacine Adenovirus Identified during an Outbreak of Avian Chlamydiosis and Human Psittacosis: Zoonosis Associated with Virus-Bacterium Coinfection in Birds. PLoS Negl. Trop. Dis. 2014, 8, e3318. [Google Scholar] [CrossRef] [PubMed]
- Benkő, M.; Harrach, B.; Kremer, E.J. Do nonhuman primate or bat adenoviruses pose a risk for human health? Future Microbiol. 2014, 9, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-Y.; Halder, S.; Agbandje-McKenna, M. Parvovirus glycan interactions. Curr. Opin. Virol. 2014, 7, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zeng, W.; Zhang, X.; Li, S. The genetic evolution of canine parvovirus—A new perspective. PLoS ONE 2017, 12, e0175035. [Google Scholar] [CrossRef]
- Simon, M.A. Simian parvoviruses: Biology and implications for research. Comp. Med. 2008, 58, 47–50. [Google Scholar]
- Brown, K.E.; Liu, Z.; Gallinella, G.; Wong, S.; Mills, I.P.; O’Sullivan, M.G. Simian Parvovirus Infection: A Potential Zoonosis. J. Infect. Dis. 2004, 190, 1900–1907. [Google Scholar] [CrossRef]
- Dugan, A.S.; Gasparovic, M.L.; Atwood, W.J. Direct Correlation between Sialic Acid Binding and Infection of Cells by Two Human Polyomaviruses (JC Virus and BK Virus). J. Virol. 2008, 82, 2560–2564. [Google Scholar] [CrossRef]
- Liu, C.K.; Wei, G.; Atwood, W.J. Infection of Glial Cells by the Human Polyomavirus JC Is Mediated by an N-Linked Glycoprotein Containing Terminal α(2-6)-Linked Sialic Acids. J. Virol. 1998, 72, 4643–4649. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.M.; Liu, Y.; Neu, U.; Gilbert, M.; Ehlers, B.; Feizi, T.; Stehle, T. Crystallographic and Glycan Microarray Analysis of Human Polyomavirus 9 VP1 Identifies N-Glycolyl Neuraminic Acid as a Receptor Candidate. J. Virol. 2014, 88, 6100–6111. [Google Scholar] [CrossRef] [PubMed]
- Scuda, N.; Madinda, N.F.; Akoua-Koffi, C.; Adjogoua, E.V.; Wevers, D.; Hofmann, J.; Cameron, K.N.; Leendertz, S.A.J.; Couacy-Hymann, E.; Robbins, M.; et al. Novel Polyomaviruses of Nonhuman Primates: Genetic and Serological Predictors for the Existence of Multiple Unknown Polyomaviruses within the Human Population. PLoS Pathog. 2013, 9, e1003429. [Google Scholar] [CrossRef]
- Parry, J.V.; Gardner, S.D. Human exposure to bovine polyomavirus: A zoonosis? Arch. Virol. 1986, 87, 287–296. [Google Scholar] [CrossRef]
- Alexander, K.A.; Carlson, C.J.; Lewis, B.L.; Getz, W.M.; Marathe, M.V.; Eubank, S.G.; Sanderson, C.E.; Blackburn, J.K. The Ecology of Pathogen Spillover and Disease Emergence at the Human-Wildlife-Environment Interface. In The Connections between Ecology and Infectious Disease; Hurst, C.J., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 267–298. [Google Scholar] [CrossRef]
- Ryu, W.-S. New Emerging Viruses. Mol. Virol. Hum. Pathog. Viruses 2017. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef] [PubMed]
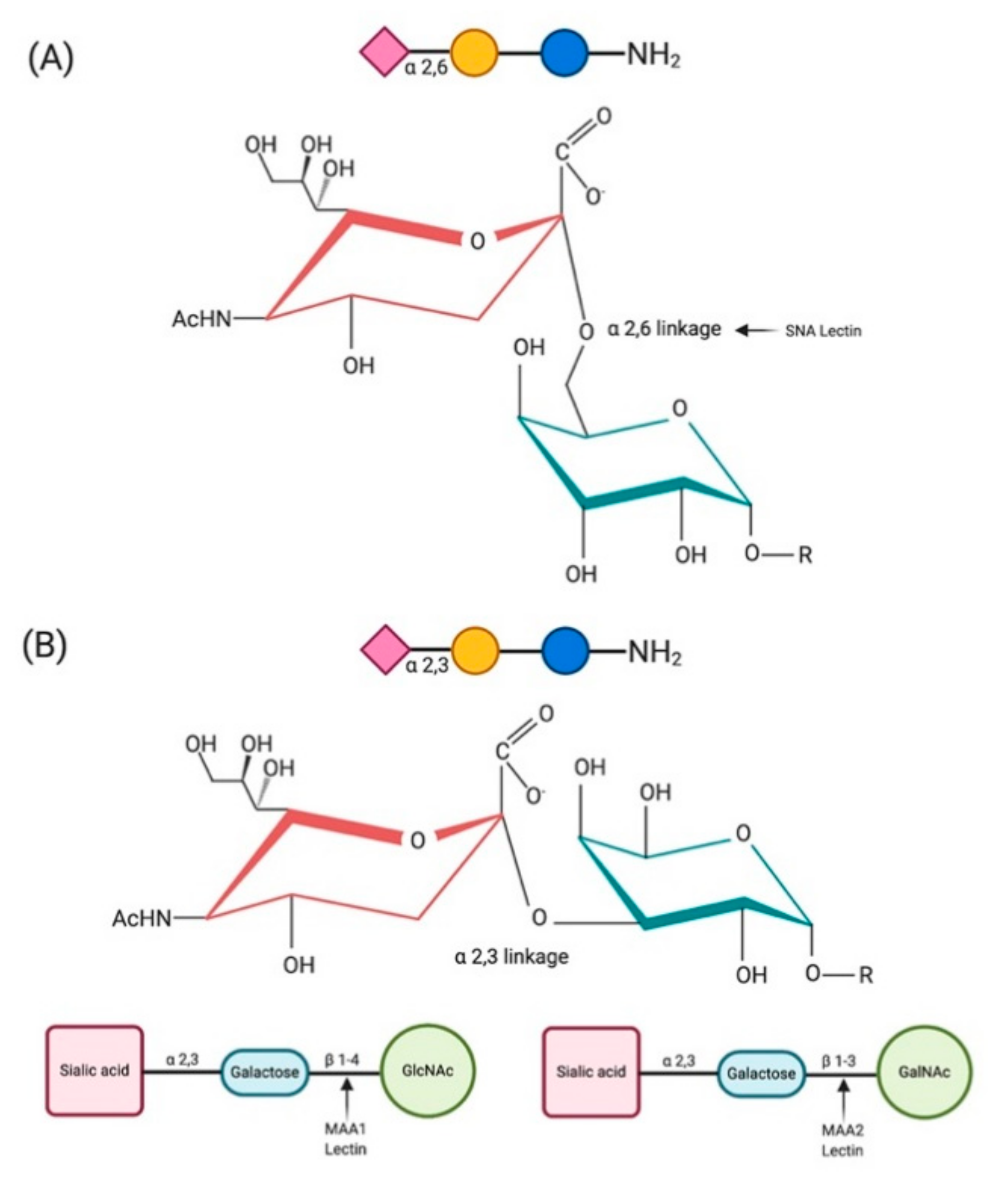
| Host | Distribution of SA α2,6-Gal | Distribution of SA α2,3-Gal | References |
|---|---|---|---|
| Humans | Ciliated and non-ciliated cells in respiratory tract; ileal epithelium | Ciliated cells in bronchioles and alveoli; colon epithelium; endothelial cells of blood vessels and inflammatory cells | [22,30,36,38,39,40,41,56] |
| Non-human primates | Goblet cells of submucosal glands and submucosal connective tissue; ciliated cells on epithelium in URT | Goblet cells of submucosal glands and submucosal connective tissue; Type II pneumocytes in lungs | [22,37,41,42] |
| Swine | Ciliated epithelia and goblet cells in trachea and bronchus; alveolar epithelium; duodenum; colon | Alveolar epithelium; duodenum; colon | [31,43,44,45] |
| Equines | Along nasal mucosa to bronchus; goblet cells | Ciliated nasal mucosa; trachea; bronchus; goblet cells | [49] |
| Bovines | Deficient in trachea; | Trachea | [45,47,48,49] |
| Camelidae | Not reported | Nasal respiratory epithelium; alveolar epithelial cells | [50] |
| Canines | Goblet cells and sub-epithelial regions of nasal mucosa and trachea; lamina propria; large intestine | Nasal mucosa; trachea; bronchi; alveoli; goblet cells and sub-epithelial regions of nasal mucosa and trachea; large intestine | [51,52] |
| Felines | Ciliated pseudostratified epithelial cells; goblet cells; alveolar epithelial cells | Ciliated pseudostratified epithelial cells; goblet cells; sub-epithelial connective tissue; alveolar epithelial cells | [51,53] |
| Bats | Lamina propria; submucosa of trachea and bronchi; alveoli; goblet cells; serosa of intestine | Epithelial cells of trachea; mucosal lining of intestinal villi | [54,55] |
| Plateau Pika | Lamina propria and mucous glands of trachea; alveolar epithelial cells; duodenum; ileum; rectum | Lamina propria and mucous glands of trachea; alveolar epithelial cells; duodenum; ileum; rectum. | [57] |
| Raccoon | Epithelium and sub-epithelial regions in URT | Epithelial and sub-epithelial regions of trachea and bronchi. | [58] |
| Guinea pig | Epithelial cells of nasal and tracheal mucosa | Alveolar cells and vascular endothelial cells. | [59] |
| Ferrets | Ciliated cells and submucosal glands in trachea and bronchus; alveoli | Lamina propria and sub-mucosal areas; alveoli | [53,60,61] |
| Hamsters | Distal end of the nasal cavity; pharynx; trachea; bronchus | Proximal end of the nasal cavity; pharynx; trachea; bronchus; lungs | [62,63] |
| Mice | Sub-epithelial locations; trachea; bronchi; bronchioles; alveolar cells; intestines; lymphoid nodules of cecum | Sub-epithelial locations; trachea; bronchi; bronchioles; alveolar cells; intestines | [64] |
| Galliformes | Nasal cavity to lungs; tracheal epithelium of chickens and quails | Nasal cavity to lungs; trachea of ducks; intestines | [29,65,66] |
| Passeriformes | Epithelial cells of respiratory tract; sub-epithelial regions and glands | Epithelial cells of respiratory tract (predominant); sub-epithelial regions and glands; intestines | [21,66] |
| Columbiformes | Respiratory tract | Respiratory tract | |
| Anseriformes | Respiratory and intestinal tracts; lungs; ileal and cecal enterocytes; duodenum; jejunum | Respiratory and intestinal tracts; respiratory epithelial cells; ciliated cells; goblet cells; sub-epithelial regions of trachea and bronchi; goblet cells of small and large intestines | [21,66] |
| Charadriiformes | Respiratory tract epithelium; marginal in intestines | Respiratory tract epithelium | [21] |
| Gruiformes, Pelecaniformes, Gaviiformes, Ciconiiformes | Respiratory epithelium | Respiratory and intestinal epithelium | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuchipudi, S.V.; Nelli, R.K.; Gontu, A.; Satyakumar, R.; Surendran Nair, M.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. https://doi.org/10.3390/v13020262
Kuchipudi SV, Nelli RK, Gontu A, Satyakumar R, Surendran Nair M, Subbiah M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses. 2021; 13(2):262. https://doi.org/10.3390/v13020262
Chicago/Turabian StyleKuchipudi, Suresh V, Rahul K Nelli, Abhinay Gontu, Rashmi Satyakumar, Meera Surendran Nair, and Murugan Subbiah. 2021. "Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover" Viruses 13, no. 2: 262. https://doi.org/10.3390/v13020262
APA StyleKuchipudi, S. V., Nelli, R. K., Gontu, A., Satyakumar, R., Surendran Nair, M., & Subbiah, M. (2021). Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses, 13(2), 262. https://doi.org/10.3390/v13020262







