RNA Viruses in Aquatic Unicellular Eukaryotes
Abstract
1. Introduction
2. Marine RNA Viruses
3. Protist Virus Characterization
4. Taxonomy of Protist Viruses
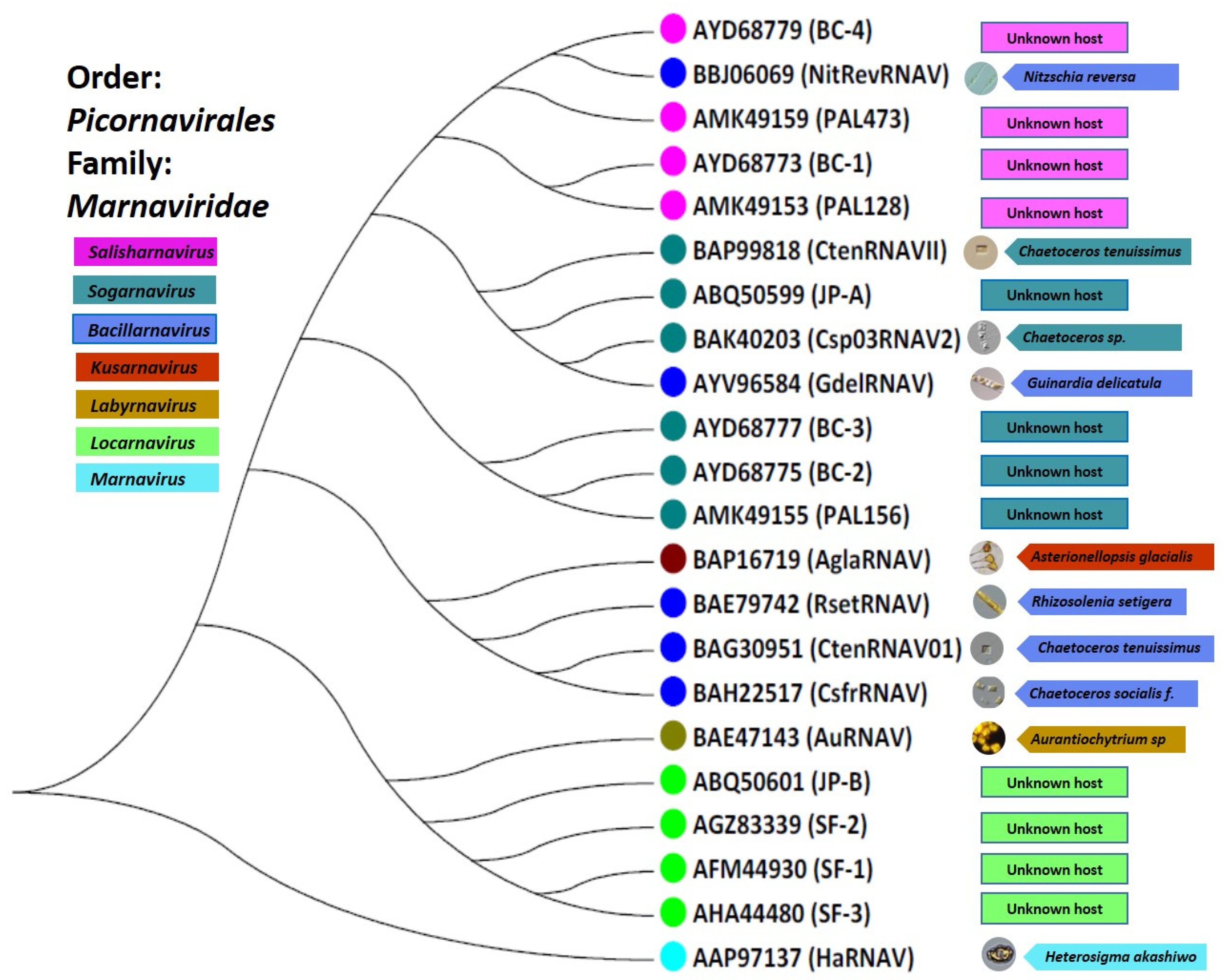
4.1. Picornaviridae in Protists
| Protist Viruses in the Order Picornavirales | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genus | Species/Virus | Virus Name Abbreviation | Genome | Host or Source | Country | References | ||
| Size (nt) | ORFs 1 | |||||||
| Marnavirus | Heterosigma akashiwo RNA virus | HaRNAV | 8587 | One | Raphidophyte | Heterosigma akashiwo | Canada | Lang et al., 2004; Tai et al., 2003 [18,23] |
| Labyrnavirus | Aurantiochytrium ssRNA virus | AuRNAV01 | 9035 | Two | Thraustochytrids | Aurantiochytrium sp. | Japan | Takao et al., 2006 [75] |
| Locarnavirus | Marine RNA virus JP-B | JP-B | 8926 | Two | Unknown | Coastal marine | Canada | Culley et al., 2007 [70] |
| Marine RNA virus SF-2 | SF-2 | 9321 | Two | Unknown | Coastal wastewater | USA | Greninger and DeRisi, 2015 [27] | |
| Marine RNA virus SF-1 | SF-1 | 8970 | Two | Unknown | Coastal wastewater | USA | Greninger and DeRisi, 2015 [27] | |
| Marine RNA virus SF-3 | SF-3 | 8648 | One | Unknown | Coastal wastewater | USA | Greninger and DeRisi, 2015 [27] | |
| Kusarnavirus | Asterionellopsis glacialis RNA virus | AglaRNAV | 8842 | Two | Diatom | Asterionellopsis glacialis | Japan | Tomaru et al., 2012 [63] |
| Bacillarnavirus | Chaetoceros tenuissimus RNA virus 01 | CtenRNAV Type 1 | 9431 | Two | Diatom | Chaetoceros tenuissimus | Japan | Shirai et al., 2008 [76] |
| Rhizosolenia setigera RNA virus | RsetRNAV | 8877 | Two | Diatom | Rhizosolenia setigera | Japan | Nagasaki et al., 2004 [22] | |
| Chaetoceros socialis f. radians RNA virus 01 | CsfrRNAV | 9467 | Two | Diatom | Chaetoceros socialis f. radians | Japan | Tomaru et al., 2009 [62] | |
| Guinardia delicatula RNA virus | GdelRNAV | 9233 | Two | Diatom | Guinardia delicatula | France | Arsenieff et al., 2018 [60] | |
| Nitzschia reversa RNA virus | NitRevRNAV | ~9000 | Two | Diatom | Nitzschia reversa | Japan | Toyoda et al., 2019 [77] | |
| Salisharnavirus | Marine RNA virus BC-4 | BC-4 | 8593 | Two | Unknown | Coastal/oceanic marine | Canada | Vlok et al., 2019b [47] |
| Marine RNA virus PAL473 | PAL473 | 6360 | Two | Unknown | Coastal marine | USA | Miranda et al., 2016 [71] | |
| Marine RNA virus BC-1 | BC-1 | 8638 | Two | Unknown | Coastal marine | Canada | Vlok et al., 2019b [47] | |
| Marine RNA virus PAL128 | PAL128 | 8660 | Two | Unknown | Coastal marine | USA | Miranda et al., 2016 [71] | |
| Sogarnavirus | Marine RNA virus BC-2 | BC-2 | 8843 | Two | Unknown | Coastal marine | Canada | Vlok et al., 2019b [47] |
| Marine RNA virus PAL156 | PAL156 | 7897 | Two | Unknown | Coastal marine | USA | Miranda et al., 2016 [71] | |
| Marine RNA virus BC-3 | BC-3 | 8496 | Two | Unknown | Coastal marine | Canada | Vlok et al., 2019b [47] | |
| Marine RNA virus JP-A | JP-A | 9236 | Two | Unknown | Coastal marine | Canada | Culley et al., 2007 [70] | |
| Chaetoceros tenuissimus RNA virus type II | CtenRNAV Type 2 | 9562 | Two | Diatom | Chaetoceros tenuissimus | Japan | Kimura and Tomarua, 2015 [78] | |
| Chaetoceros species RNA virus01 | Csp03RNAV | 9417 | Two | Diatom | Chaetoceros sp. | Japan | Tomaru et al., 2013 [64] | |
4.2. Alvernaviridae in Protists
4.3. Reoviridae in Protists
| Protist Viruses in the Family Alvernaviridae | ||||||||
| Genus | Species | Virus Name Abbreviation | Genome | Host or Source | Country | References | ||
| Size (nt) | ORFs 1 | |||||||
| Dinornavirus | Heterocapsa circularisquamaRNA virus 01 | HcRNAV | 4400 | Two | Dinoflagellate | Heterocapsa circularisquama | Japan | Nagasaki et al., 2005a; Tomaru et al., 2004 [21,24] |
| Protist Viruses in Family Reoviridae | ||||||||
| Genus | Species | Virus Name Abbreviation | Genome | Host or Source | Country | References | ||
| Size (nt) | ORFs | |||||||
| Mimoreovirus | Micromonas pusillareovirus | MpRNAV-01B | 4400 | 11 | Prasinophyceae | Microalga Micromonas pusilla | France | Brussaard et al., 2004 [25] |
5. Host Specificity of Protist Viruses
6. RNA Viruses of Diatoms
6.1. Diatoms of the Genus Rhizosolenia
6.2. Diatoms of the Genus Chaetoceros
6.3. Diatom of the Genus Asterionella
6.4. Diatom of the Genus Guinardia
6.5. Diatoms of the Genus Nitzschia
7. Viruses of the Family Raphidophyceae
8. Viruses of the Family Thraustochytriaceae
9. Viruses of Dinoflagellates
10. Viruses of the Family Prasinophyceae
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Javaux, E.J.; Knoll, A.H.; Walter, M.R. Morphological and ecological complexity in early eukaryotic ecosystems. Nature 2001, 412, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M. Molecular classification and identification of toxic dinoflagellate, Alexandrium. Nippon Suisan Gakk 1998, 64, 583–587. [Google Scholar] [CrossRef]
- Park, J.S.; Simpson, A.G. Diversity of Heterotrophic Protists from Extremely Hypersaline Habitats. Protist 2015, 166, 422–437. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukes, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef]
- James, T.Y.; Pelin, A.; Bonen, L.; Ahrendt, S.; Sain, D.; Corradi, N.; Stajich, J.E. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr. Biol. 2013, 23, 1548–1553. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Salamon, P.; Andresen, B.; Mahaffy, J.M.; Segall, A.M.; Mead, D.; Azam, F.; Rohwer, F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 2002, 99, 14250–14255. [Google Scholar] [CrossRef]
- Djamali, E.; Nulton, J.D.; Turner, P.J.; Rohwer, F.; Salamon, P. Heat output by marine microbial and viral communities. J. Non-Equil. Thermody 2012, 37, 291–313. [Google Scholar] [CrossRef][Green Version]
- Lee, R.E. Systemic viral material in the cells of the freshwater red alga Sirodotia tenuissima (Holden) skuja. J. Cell Sci. 1971, 8, 623–631. [Google Scholar] [PubMed]
- Van Etten, J.L.; Lane, L.C.; Meints, R.H. Viruses and viruslike particles of eukaryotic algae. Microbiol. Rev. 1991, 55, 586–620. [Google Scholar] [CrossRef]
- Van Etten, J.L.; Meints, R.H.; Burbank, D.E.; Kuczmarski, D.; Cuppels, D.A.; Lane, L.C. Isolation and characterization of a virus from the intracellular green alga symbiotic with Hydra viridis. Virology 1981, 113, 704–711. [Google Scholar] [CrossRef]
- Kristensen, D.M.; Mushegian, A.R.; Dolja, V.V.; Koonin, E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010, 18, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.F.; Shi, M.; Eden, J.S.; Zhang, Y.Z.; Holmes, E.C. Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics. Viruses 2019, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Thurber, R.V. Viruses manipulate the marine environment. Nature 2009, 459, 207–212. [Google Scholar] [CrossRef]
- Tai, V.; Lawrence, J.E.; Lang, A.S.; Chan, A.M.; Culley, A.I.; Suttle, C.A. Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 2003, 39, 343–352. [Google Scholar] [CrossRef]
- Takao, Y.; Nagasaki, K.; Mise, K.; Okuno, T.; Honda, D. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp (Thraustochytriaceae, labyrinthulea). Appl. Environ. Microbiol. 2005, 71, 4516–4522. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagasaki, K.; Tomaru, Y.; Takao, Y.; Nishida, K.; Shirai, Y.; Suzuki, H.; Nagumo, T. Previously unknown virus infects marine diatom. Appl. Environ. Microbiol. 2005, 71, 3528–3535. [Google Scholar] [CrossRef]
- Tomaru, Y.; Katanozaka, N.; Nishida, K.; Shirai, Y.; Tarutani, K.; Yamaguchi, M.; Nagasaki, K. Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat. Microb. Ecol. 2004, 34, 207–218. [Google Scholar] [CrossRef]
- Nagasaki, K.; Tomaru, Y.; Katanozaka, N.; Shirai, Y.; Nishida, K.; Itakura, S.; Yamaguchi, M. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 2004, 70, 704–711. [Google Scholar] [CrossRef]
- Lang, A.S.; Culley, A.I.; Suttle, C.A. Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology 2004, 320, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, K.; Shirai, Y.; Takao, Y.; Mizumoto, H.; Nishida, K.; Tomaru, Y. Comparison of genome sequences of single-stranded RNA viruses infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 2005, 71, 8888–8894. [Google Scholar] [CrossRef]
- Brussaard, C.P.; Noordeloos, A.A.; Sandaa, R.A.; Heldal, M.; Bratbak, G. Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology 2004, 319, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Gustavsen, J.A.; Winget, D.M.; Tian, X.; Suttle, C.A. High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities. Front. Microbiol. 2014, 5, 703. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; DeRisi, J.L. Draft Genome Sequences of Marine RNA Viruses SF-1, SF-2, and SF-3 Recovered from San Francisco Wastewater. Genome Announc. 2015, 3, e00653-15. [Google Scholar] [CrossRef]
- Vlok, M.; Lang, A.S.; Suttle, C.A. Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales. Virus Evol. 2019, 5, 56. [Google Scholar] [CrossRef]
- Steward, G.F.; Culley, A.I.; Mueller, J.A.; Wood-Charlson, E.M.; Belcaid, M.; Poisson, G. Are we missing half of the viruses in the ocean? ISME J. 2013, 7, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Rise, M.L.; Culley, A.I.; Steward, G.F. RNA viruses in the sea. FEMS Microbiol. Rev. 2009, 33, 295–323. [Google Scholar] [CrossRef]
- Tomaru, Y.; Fujii, N.; Oda, S.; Toyoda, K.; Nagasaki, K. Dynamics of diatom viruses on the western coast of Japan. Aquat. Microb. Ecol. 2011, 63, 223–230. [Google Scholar] [CrossRef]
- Tomaru, Y.; Hata, N.; Masuda, T.; Tsuji, M.; Igata, K.; Masuda, Y.; Yamatogi, T.; Sakaguchi, M.; Nagasaki, K. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 2007, 9, 1376–1383. [Google Scholar] [CrossRef]
- Culley, A.I.; Lang, A.S.; Suttle, C.A. High diversity of unknown picorna-like viruses in the sea. Nature 2003, 424, 1054–1057. [Google Scholar] [CrossRef] [PubMed]
- Culley, A.I.; Steward, G.F. New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl. Environ. Microbiol. 2007, 73, 5937–5944. [Google Scholar] [CrossRef] [PubMed]
- Culley, A.I.; Lang, A.S.; Suttle, C.A. Metagenomic analysis of coastal RNA virus communities. Science 2006, 312, 1795–1798. [Google Scholar] [CrossRef]
- Urayama, S.; Takaki, Y.; Nishi, S.; Yoshida-Takashima, Y.; Deguchi, S.; Takai, K.; Nunoura, T. Unveiling the RNA virosphere associated with marine microorganisms. Mol. Ecol. Resour. 2018, 18, 1444–1455. [Google Scholar] [CrossRef]
- Sadeghi, M.; Popov, V.; Guzman, H.; Phan, T.G.; Vasilakis, N.; Tesh, R.; Delwart, E. Genomes of viral isolates derived from different mosquitos species. Virus Res. 2017, 242, 49–57. [Google Scholar] [CrossRef]
- Sadeghi, M.; Kapusinszky, B.; Yugo, D.M.; Phan, T.G.; Deng, X.T.; Kanevsky, I.; Opriessnig, T.; Woolums, A.R.; Hurley, D.J.; Meng, X.J.; et al. Virome of US bovine calf serum. Biologicals 2017, 46, 64–67. [Google Scholar] [CrossRef]
- Sadeghi, M.; Altan, E.; Deng, X.T.; Barker, C.M.; Fang, Y.; Coffey, L.L.; Delwart, E. Virome of > 12 thousand Culex mosquitoes from throughout California. Virology 2018, 523, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Delwart, E.L. Viral metagenomics. Rev. Med. Virol. 2007, 17, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Bench, S.R.; Hanson, T.E.; Williamson, K.E.; Ghosh, D.; Radosovich, M.; Wang, K.; Wommack, K.E. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 2007, 73, 7629–7641. [Google Scholar] [CrossRef] [PubMed]
- Wolf, Y.I.; Silas, S.; Wang, Y.; Wu, S.; Bocek, M.; Kazlauskas, D.; Krupovic, M.; Fire, A.; Dolja, V.V.; Koonin, E.V. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 2020, 5, 1262–1270. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Chen, Y.M.; Wang, W.; Qin, X.C.; Holmes, E.C. Expanding the RNA virosphere by unbiased metagenomics. Annu. Virol. 2019, 6, 119–139. [Google Scholar] [CrossRef]
- Dolja, V.V.; Koonin, E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018, 244, 36–52. [Google Scholar] [CrossRef]
- Culley, A.I.; Mueller, J.A.; Belcaid, M.; Wood-Charlson, E.M.; Poisson, G.; Steward, G.F. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio 2014, 5, e01210-14. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef] [PubMed]
- Vlok, M.; Lang, A.S.; Suttle, C.A. Marine RNA Virus Quasispecies Are Distributed throughout the Oceans. mSphere 2019, 4, e00157-19. [Google Scholar] [CrossRef] [PubMed]
- Yau, S.; Seth-Pasricha, M. Viruses of Polar Aquatic Environments. Viruses 2019, 11, 189. [Google Scholar] [CrossRef]
- Munke, A.; Kimura, K.; Tomaru, Y.; Okamoto, K. Capsid Structure of a Marine Algal Virus of the Order Picornavirales. J. Virol. 2020, 94, e01855-19. [Google Scholar] [CrossRef]
- Greninger, A.L. A decade of RNA virus metagenomics is (not) enough. Virus Res. 2018, 244, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Tomaru, Y.; Kimura, K. Novel Protocol for Estimating Viruses Specifically Infecting the Marine Planktonic Diatoms. Diversity 2020, 12, 225. [Google Scholar] [CrossRef]
- Suttle, C.A.; Chan, A.M.; Cottrell, M.T. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 1991, 57, 721–726. [Google Scholar] [CrossRef]
- Bergh, O.; Borsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, e368. [Google Scholar] [CrossRef]
- Bonami, J.R.; Zhang, S. Viral diseases in commercially exploited crabs: A review. J. Invertebr. Pathol. 2011, 106, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, H.F.; Heuer, O.E.; Lorenzen, N.; Otte, L.; Olesen, N.J. Isolation of viral haemorrhagic septicaemia virus (VHSV) from wild marine fish species in the Baltic Sea, Kattegat, Skagerrak and the North Sea. Virus Res. 1999, 63, 95–106. [Google Scholar] [CrossRef]
- Jensen, T.; van de Bildt, M.; Dietz, H.H.; Andersen, T.H.; Hammer, A.S.; Kuiken, T.; Osterhaus, A. Another phocine distemper outbreak in Europe. Science 2002, 297, 209. [Google Scholar] [CrossRef]
- Renault, T.; Novoa, B. Viruses infecting bivalve molluscs. Aquat. Living Resour. 2004, 17, 397–409. [Google Scholar] [CrossRef]
- Levin, R.A.; Voolstra, C.R.; Weynberg, K.D.; van Oppen, M.J. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 2017, 11, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Arsenieff, L.; Simon, N.; Rigaut-Jalabert, F.; Le Gall, F.; Chaffron, S.; Corre, E.; Com, E.; Bigeard, E.; Baudoux, A.C. First Viruses Infecting the Marine Diatom Guinardia delicatula. Front. Microbiol. 2018, 9, 3235. [Google Scholar] [CrossRef] [PubMed]
- Tomaru, Y.; Shirai, Y.; Suzuki, H.; Nagumo, T.; Nagasaki, K. Isolation and characterization of a new single-stranded DNA virus infecting the cosmopolitan marine diatom Chaetoceros dehilis. Aquat. Microb. Ecol. 2008, 50, 103–112. [Google Scholar] [CrossRef]
- Tomaru, Y.; Takao, Y.; Suzuki, H.; Nagumo, T.; Nagasaki, K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 2009, 75, 2375–2381. [Google Scholar] [CrossRef]
- Tomaru, Y.; Toyoda, K.; Kimura, K.; Hata, N.; Yoshida, M.; Nagasaki, K. First evidence for the existence of pennate diatom viruses. ISME J. 2012, 6, 1445–1448. [Google Scholar] [CrossRef]
- Tomaru, Y.; Toyoda, K.; Kimura, K.; Takao, Y.; Sakurada, K.; Nakayama, N.; Nagasaki, K. Isolation and characterization of a single-stranded RNA virus that infects the marine planktonic diatom Chaetoceros sp (SS08-C03). Phycol. Res. 2013, 61, 27–36. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Wolf, Y.I.; Krupovic, M.; Zhang, Y.Z.; Maes, P.; Dolja, V.V.; Koonin, E.V. Classify viruses—The gain is worth the pain. Nature 2019, 566, 318–320. [Google Scholar] [CrossRef] [PubMed]
- Poch, O.; Blumberg, B.M.; Bougueleret, L.; Tordo, N. Sequence Comparison of 5 Polymerases (L-Proteins) of Unsegmented Negative-Strand Rna Viruses—Theoretical Assignment of Functional Domains. J. Gen. Virol. 1990, 71, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Vieth, S.; Torda, A.E.; Asper, M.; Schmitz, H.; Gunther, S. Sequence analysis of L RNA of Lassa virus. Virology 2004, 318, 153–168. [Google Scholar] [CrossRef]
- Jacome, R.; Becerra, A.; de Leon, S.P.; Lazcano, A. Structural Analysis of Monomeric RNA-Dependent Polymerases: Evolutionary and Therapeutic Implications. PLoS ONE 2015, 10, e0139001. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–545. [Google Scholar] [CrossRef]
- Culley, A.I.; Lang, A.S.; Suttle, C.A. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 2007, 4, 69. [Google Scholar] [CrossRef]
- Miranda, J.A.; Culley, A.I.; Schvarcz, C.R.; Steward, G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016, 18, 3714–3727. [Google Scholar] [CrossRef]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008, 153, 715–727. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucia-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and Evolution of the Global RNA Virome. mBio 2018, 9, e02329-18. [Google Scholar] [CrossRef]
- Takao, Y.; Mise, K.; Nagasaki, K.; Okuno, T.; Honda, D. Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp. J. Gen. Virol. 2006, 87, 723–733. [Google Scholar] [CrossRef]
- Shirai, Y.; Tomaru, Y.; Takao, Y.; Suzuki, H.; Nagumo, T.; Nagasaki, K. Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 2008, 74, 4022–4027. [Google Scholar] [CrossRef]
- Toyoda, K.; Kimura, K.; Osada, K.; Williams, D.M.; Adachi, T.; Yamada, K.; Tomaru, Y. Novel marine diatom ssRNA virus NitRevRNAV infecting Nitzschia reversa. Plant Ecol. Evol. 2019, 152, 178–187. [Google Scholar] [CrossRef]
- Kimura, K.; Tomarua, Y. Discovery of Two Novel Viruses Expands the Diversity of Single-Stranded DNA and Single-Stranded RNA Viruses Infecting a Cosmopolitan Marine Diatom. Appl. Environ. Microb. 2015, 81, 1120–1131. [Google Scholar] [CrossRef]
- Somera, M.; Sarmiento, C.; Truve, E. Overview on Sobemoviruses and a Proposal for the Creation of the Family Sobemoviridae. Viruses 2015, 7, 3076–3115. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Carstens, E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch. Virol. 2012, 157, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Short, S.M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 2012, 14, 2253–2271. [Google Scholar] [CrossRef] [PubMed]
- Tarutani, K.; Nagasaki, K.; Yamaguchi, M. Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton Heterosigma akashiwo. Appl. Environ. Microbiol. 2000, 66, 4916–4920. [Google Scholar] [CrossRef]
- Mizumoto, H.; Tomaru, Y.; Takao, Y.; Shirai, Y.; Nagasaki, K. Diverse responses of the bivalve-killing dinoflagellate Heterocapsa circularisquama to infection by a single-stranded RNA virus. Appl. Environ. Microbiol. 2008, 74, 3105–3111. [Google Scholar] [CrossRef]
- Kimura, K.; Tomaru, Y. Effects of temperature and salinity on diatom cell lysis by DNA and RNA viruses. Aquat. Microb. Ecol. 2017, 79, 79–83. [Google Scholar] [CrossRef]
- Tomaru, Y.; Toyoda, K.; Kensuke, K.K. Marine diatom viruses and their hosts: Resistance mechanisms and population dynamics. Perspect. Phycol. 2015, 2, 69–81. [Google Scholar] [CrossRef]
- Tarutani, K.; Nagasaki, K.; Yamaguchi, M. Virus adsorption process determines virus susceptibility in Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 2006, 42, 209–213. [Google Scholar] [CrossRef]
- Mizumoto, H.; Tomaru, Y.; Takao, Y.; Shirai, Y.; Nagasaki, K. Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection. J. Virol. 2007, 81, 1372–1378. [Google Scholar] [CrossRef] [PubMed]
- Fricks, C.E.; Hogle, J.M. Cell-induced conformational change in poliovirus: Externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 1990, 64, 1934–1945. [Google Scholar] [CrossRef]
- Rossmann, M.G.; He, Y.N.; Kuhn, R.J. Picornavirus-receptor interactions. Trends Microbiol. 2002, 10, 324–331. [Google Scholar] [CrossRef]
- Pierella Karlusich, J.J.; Ibarbalz, F.M.; Bowler, C. Phytoplankton in the Tara Ocean. Ann. Rev. Mar. Sci. 2020, 12, 233–265. [Google Scholar] [CrossRef] [PubMed]
- Malviya, S.; Scalco, E.; Audic, S.; Vincenta, F.; Veluchamy, A.; Poulain, J.; Wincker, P.; Iudicone, D.; de Vargas, C.; Bittner, L.; et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 2016, 113, E1516–E1525. [Google Scholar] [CrossRef] [PubMed]
- Follows, M.J.; Dutkiewicz, S.; Grant, S.; Chisholm, S.W. Emergent biogeography of microbial communities in a model ocean. Science 2007, 315, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Smetacek, V. Diatoms and the ocean carbon cycle. Protist 1999, 150, 25–32. [Google Scholar] [CrossRef]
- Smetacek, V. Seeing is Believing: Diatoms and the Ocean Carbon Cycle Revisited. Protist 2018, 169, 791–802. [Google Scholar] [CrossRef]
- Proctor, L.M.; Fuhrman, J.A. Roles of Viral-Infection in Organic Particle-Flux. Mar. Ecol. Prog. Ser. 1991, 69, 133–142. [Google Scholar] [CrossRef]
- Raven, J.A.; Waite, A.M. The evolution of silicification in diatoms: Inescapable sinking and sinking as escape? New Phytol. 2004, 162, 45–61. [Google Scholar] [CrossRef]
- Kranzler, C.F.; Krause, J.W.; Brzezinski, M.A.; Edwards, B.R.; Biggs, W.P.; Maniscalco, M.; McCrow, J.P.; Van Mooy, B.A.S.; Bidle, K.D.; Allen, A.E.; et al. Silicon limitation facilitates virus infection and mortality of marine diatoms. Nat. Microbiol. 2019, 4, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.J. The Kingdom Protista: The Dazzling World of Living Cells By Jeremy Pickett-Heaps and Julianne Pickett-Heaps. Protist 2006, 157, 259–260. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Becerril, D.U.; Del Castillo, M.E.M. The marine planktonic diatom Rhizosolenia robusta (Bacillariophyta): Morphological studies support its transfer to a new genus, Calyptrella gen. nov. Phycologia 1996, 35, 198–203. [Google Scholar] [CrossRef]
- Shirai, Y.; Takao, Y.; Mizumoto, H.; Tomaru, Y.; Honda, D.; Magasaki, K. Genomic and phylogenetic analysis of a single-stranded RNA virus infecting Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Assoc. UK 2006, 86, 475–483. [Google Scholar] [CrossRef]
- Tas, S.; Hernandez-Becerril, D.U. Diversity and distribution of the planktonic diatom genus Chaetoceros (Bacillariophyceae) in the Golden Horn Estuary (Sea of Marmara). Diatom Res. 2017, 32, 309–323. [Google Scholar] [CrossRef]
- Kaczmarska, I.; Mather, L.; Luddington, I.A.; Muise, F.; Ehrman, J.M. Cryptic diversity in a cosmopolitan diatom known as Asterionellopsis glacialis (Fragilariaceae): Implications for ecology, biogeography, and taxonomy. Am. J. Bot. 2014, 101, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Kojadinovic-Sirinelli, M.; Villain, A.; Puppo, C.; Fon Sing, S.; Prioretti, L.; Hubert, P.; Gregori, G.; Zhang, Y.; Sassi, J.F.; Claverie, J.M.; et al. Exploring the microbiome of the “star” freshwater diatom Asterionella formosa in a laboratory context. Environ. Microbiol. 2018, 20, 3601–3615. [Google Scholar] [CrossRef]
- Gowen, R.J.; McCullough, G.; Kleppel, G.S.; Houchin, L.; Elliott, P. Are copepods important grazers of the spring phytoplankton bloom in the western Irish Sea? J. Plankton Res. 1999, 21, 465–483. [Google Scholar] [CrossRef]
- Wiltshire, K.H.; Kraberg, A.; Bartsch, I.; Boersma, M.; Franke, H.D.; Freund, J.; Gebuhr, C.; Gerdts, G.; Stockmann, K.; Wichels, A. Helgoland Roads, North Sea: 45 Years of Change. Estuar. Coast. 2010, 33, 295–310. [Google Scholar] [CrossRef]
- Guilloux, L.; Rigaut-Jalabert, F.; Jouenne, F.; Ristori, S.; Viprey, M.; Not, F.; Vaulot, D.; Simon, N. An annotated checklist of Marine Phytoplankton taxa at the SOMLIT-Astan time series off Roscoff (Western English Channel, France): Data collected from 2000 to 2010. Cah. Biol. Mar. 2013, 54, 247–256. [Google Scholar]
- Amzil, Z.; Fresnel, J.; Le Gal, D.; Billard, C. Domoic acid accumulation in French shellfish in relation to toxic species of Pseudo-nitzschia multiseries and P. pseudodelicatissima. Toxicon 2001, 39, 1245–1251. [Google Scholar] [CrossRef]
- Vesk, M.; Moestrup, O. The Flagellar Root-System in Heterosigma-Akashiwo (Raphidophyceae). Protoplasma 1987, 137, 15–28. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Nakayama, T.; Murakami, A.; Inouye, I. Phylogeny and taxonomy of the Raphidophyceae (Heterokontophyta) and Chlorinimonas sublosa gen. et sp. nov., a new marine sand-dwelling raphidophyte. J. Plant Res. 2010, 123, 333–342. [Google Scholar] [CrossRef]
- Imai, I.; Yamaguchi, M. Life cycle, physiology, ecology and red tide occurrences of the fish-killing raphidophyte Chattonella. Harmful Algae 2012, 14, 46–70. [Google Scholar] [CrossRef]
- Hiroishi, S.; Okada, H.; Imai, I.; Yoshida, T. High toxicity of the novel bloom-forming species Chattonella ovata (Raphidophyceae) to cultured fish. Harmful Algae 2005, 4, 783–787. [Google Scholar] [CrossRef]
- Nagasaki, K.; Tarutani, K.; Yamaguchi, M. Growth characteristics of Heterosigma akashiwo virus and its possible use as a microbiological agent for red tide control. Appl. Environ. Microbiol. 1999, 65, 898–902. [Google Scholar] [CrossRef]
- Lee Chang, K.J.; Nichols, C.M.; Blackburn, S.I.; Dunstan, G.A.; Koutoulis, A.; Nichols, P.D. Comparison of Thraustochytrids Aurantiochytrium sp., Schizochytrium sp., Thraustochytrium sp., and Ulkenia sp. for production of biodiesel, long-chain omega-3 oils, and exopolysaccharide. Mar Biotechnol (NY) 2014, 16, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Del Campo, J.; Keeling, P.J. Reference Tree and Environmental Sequence Diversity of Labyrinthulomycetes. J. Eukaryot. Microbiol. 2017, 64, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef]
- Marchan, L.F.; Chang, K.J.L.; Nichols, P.D.; Mitchell, W.J.; Polglase, J.L.; Gutierrez, T. Taxonomy, ecology and biotechnological applications of thraustochytrids: A review. Biotechnol. Adv. 2018, 36, 26–46. [Google Scholar] [CrossRef]
- Gomez, F. A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Syst. Biodivers. 2012, 10, 267–275. [Google Scholar] [CrossRef]
- Sournia, A. Classification and Nomenclature of Various Marine Dinoflagellates (Dinophyceae). Phycologia 1984, 23, 345–355. [Google Scholar] [CrossRef]
- Hoppenrath, M. Dinoflagellate taxonomy—A review and proposal of a revised classification. Mar. Biodivers. 2017, 47, 381–403. [Google Scholar] [CrossRef]
- Nagasaki, K.; Tomaru, Y.; Shirai, Y.; Takao, Y.; Mizumoto, H. Dinoflagellate-infecting viruses. J. Mar. Biol. Assoc. UK 2006, 86, 469–474. [Google Scholar] [CrossRef]
- Tarutani, K.; Nagasaki, K.; Itakura, S.; Yamaguchi, M. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquat. Microb. Ecol. 2001, 23, 103–111. [Google Scholar] [CrossRef]
- Tomaru, Y.; Mizumoto, H.; Nagasaki, K. Virus resistance in the toxic bloom-forming dinoflagellate Heterocapsa circularisquama to single-stranded RNA virus infection. Environ. Microbiol. 2009, 11, 2915–2923. [Google Scholar] [CrossRef]
- Lemieux, C.; Otis, C.; Turmel, M. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genom. 2014, 15, 857. [Google Scholar] [CrossRef] [PubMed]
- Kubiszyn, A.M.; Svensen, C. First record of a rare species, Polyasterias problematica (Prasinophyceae), in Balsfjord, northern Norway. Bot. Mar. 2018, 61, 421–428. [Google Scholar] [CrossRef]
- Sluiman, H.J.; Gartner, G. Taxonomic Studies on the Genus Pleurastrum (Pleurastrales, Chlorophyta). I. The Type Species, Pleurastrum-Insigne, Rediscovered and Isolated from Soil. Phycologia 1990, 29, 133–138. [Google Scholar] [CrossRef]
- Garrido, J.L.; Rodriguez, F.; Zapata, M. Occurrence of Loroxanthin, Loroxanthin Decenoate, and Loroxanthin Dodecenoate in Tetraselmis Species (Prasinophyceae, Chlorophyta)(1). J. Phycol. 2009, 45, 366–374. [Google Scholar] [CrossRef]
- Jouenne, F.; Eikrem, W.; Le Gall, F.; Marie, D.; Johnsen, G.; Vaulot, D. Prasinoderma singularis sp. nov. (Prasinophyceae, Chlorophyta), a solitary coccoid Prasinophyte from the South-East Pacific Ocean. Protist 2011, 162, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Not, F.; Latasa, M.; Marie, D.; Cariou, T.; Vaulot, D.; Simon, N. A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Appl. Environ. Microbiol. 2004, 70, 4064–4072. [Google Scholar] [CrossRef]
- Guillou, L.; Eikrem, W.; Chretiennot-Dinet, M.J.; Le Gall, F.; Massana, R.; Romari, K.; Pedros-Alio, C.; Vaulot, D. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist 2004, 155, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, H.A.; Buck, K.R. Nanoflagellates of the central California waters: Taxonomy, biogeography and abundance of primitive, green flagellates (Pedinophyceae, Prasinophyceae). Deep-Sea Res. Pt Ii 1998, 45, 1687–1707. [Google Scholar] [CrossRef]
- Filippini, M.; Middelboe, M. Viral abundance and genome size distribution in the sediment and water column of marine and freshwater ecosystems. FEMS Microbiol. Ecol. 2007, 60, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Felts, B.; Kelley, S.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 2004, 271, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, K.; Tomaru, Y.; Nakanishi, K.; Hata, N.; Katanozaka, N.; Yamaguchi, M. Dynamics of Heterocapsa circularisquama (Dinophyceae) and its viruses in Ago Bay, Japan. Aquat. Microb. Ecol. 2004, 34, 219–226. [Google Scholar] [CrossRef][Green Version]
- Ahola, T. New Phylogenetic Grouping of Positive-Sense RNA Viruses Is Concordant with Replication Complex Morphology. mBio 2019, 10, e01402-19. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadeghi, M.; Tomaru, Y.; Ahola, T. RNA Viruses in Aquatic Unicellular Eukaryotes. Viruses 2021, 13, 362. https://doi.org/10.3390/v13030362
Sadeghi M, Tomaru Y, Ahola T. RNA Viruses in Aquatic Unicellular Eukaryotes. Viruses. 2021; 13(3):362. https://doi.org/10.3390/v13030362
Chicago/Turabian StyleSadeghi, Mohammadreza, Yuji Tomaru, and Tero Ahola. 2021. "RNA Viruses in Aquatic Unicellular Eukaryotes" Viruses 13, no. 3: 362. https://doi.org/10.3390/v13030362
APA StyleSadeghi, M., Tomaru, Y., & Ahola, T. (2021). RNA Viruses in Aquatic Unicellular Eukaryotes. Viruses, 13(3), 362. https://doi.org/10.3390/v13030362






