APOBECs and Herpesviruses
Abstract
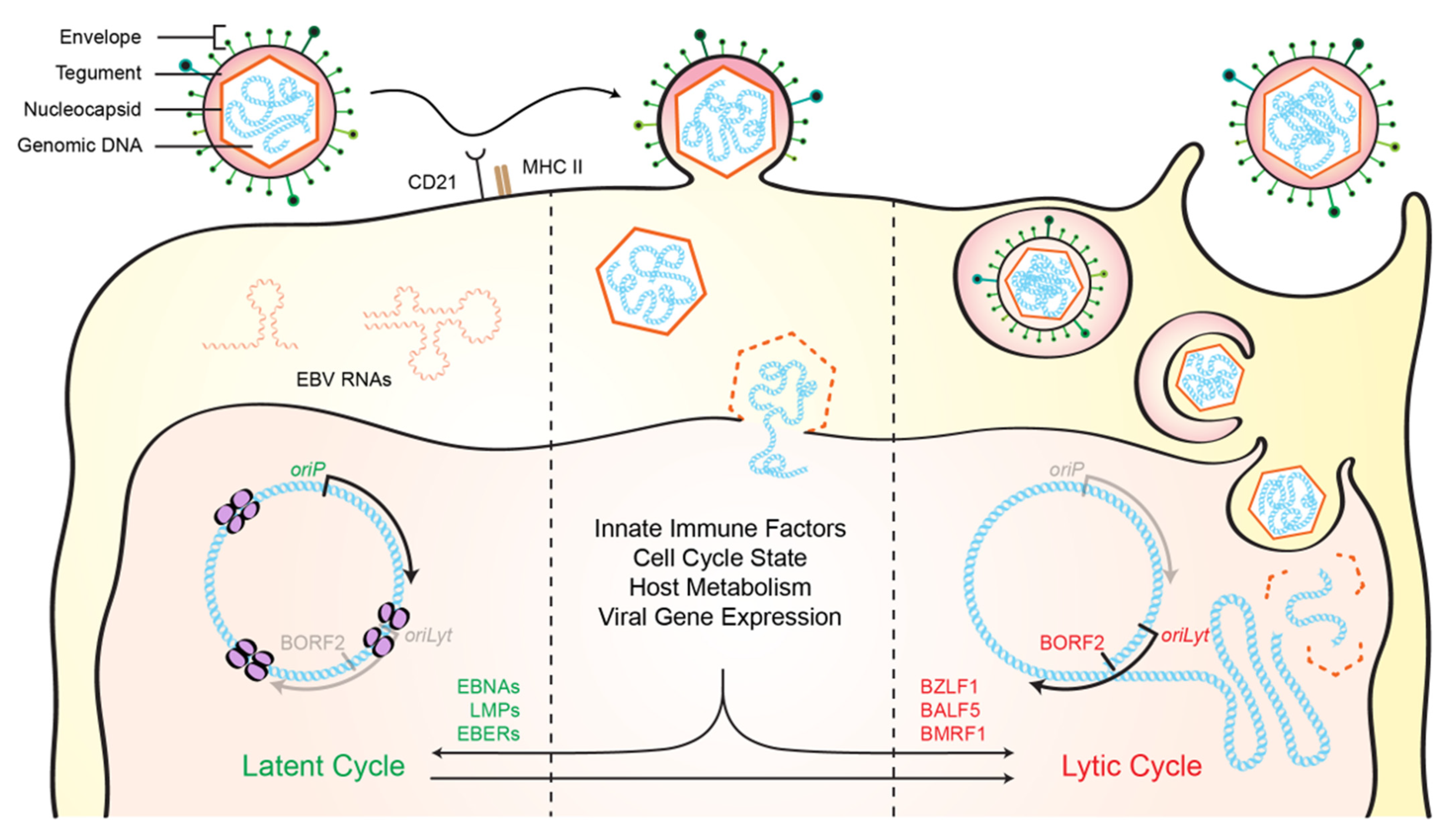
:1. Human Herpesviruses
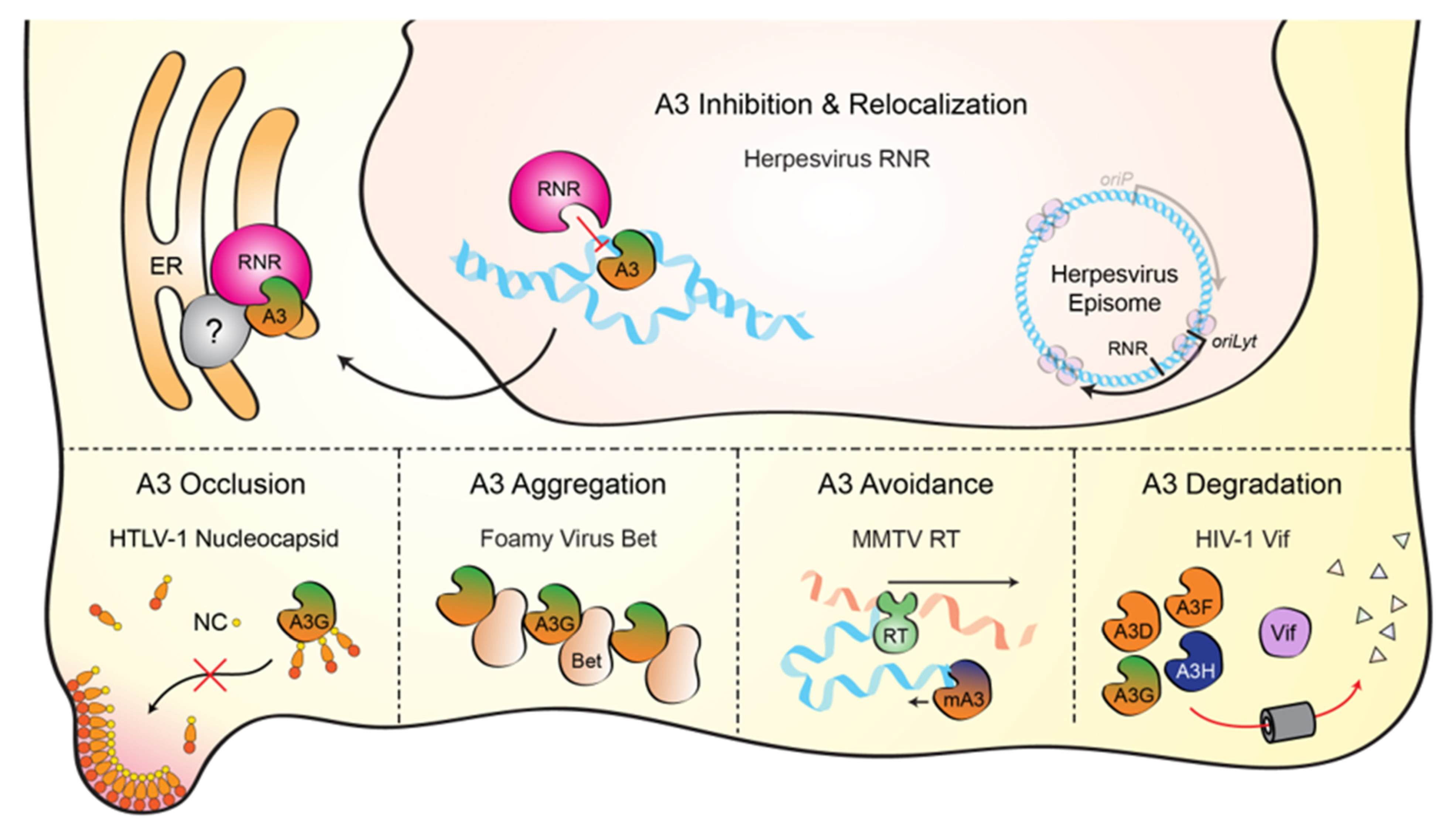
2. Discovery of a Novel Herpesvirus Ribonucleotide Reductase Interaction
3. Mechanism for EBV BORF2 Counteraction of A3B

4. Conservation of the Herpesvirus RNR-Cellular APOBEC3 Interaction
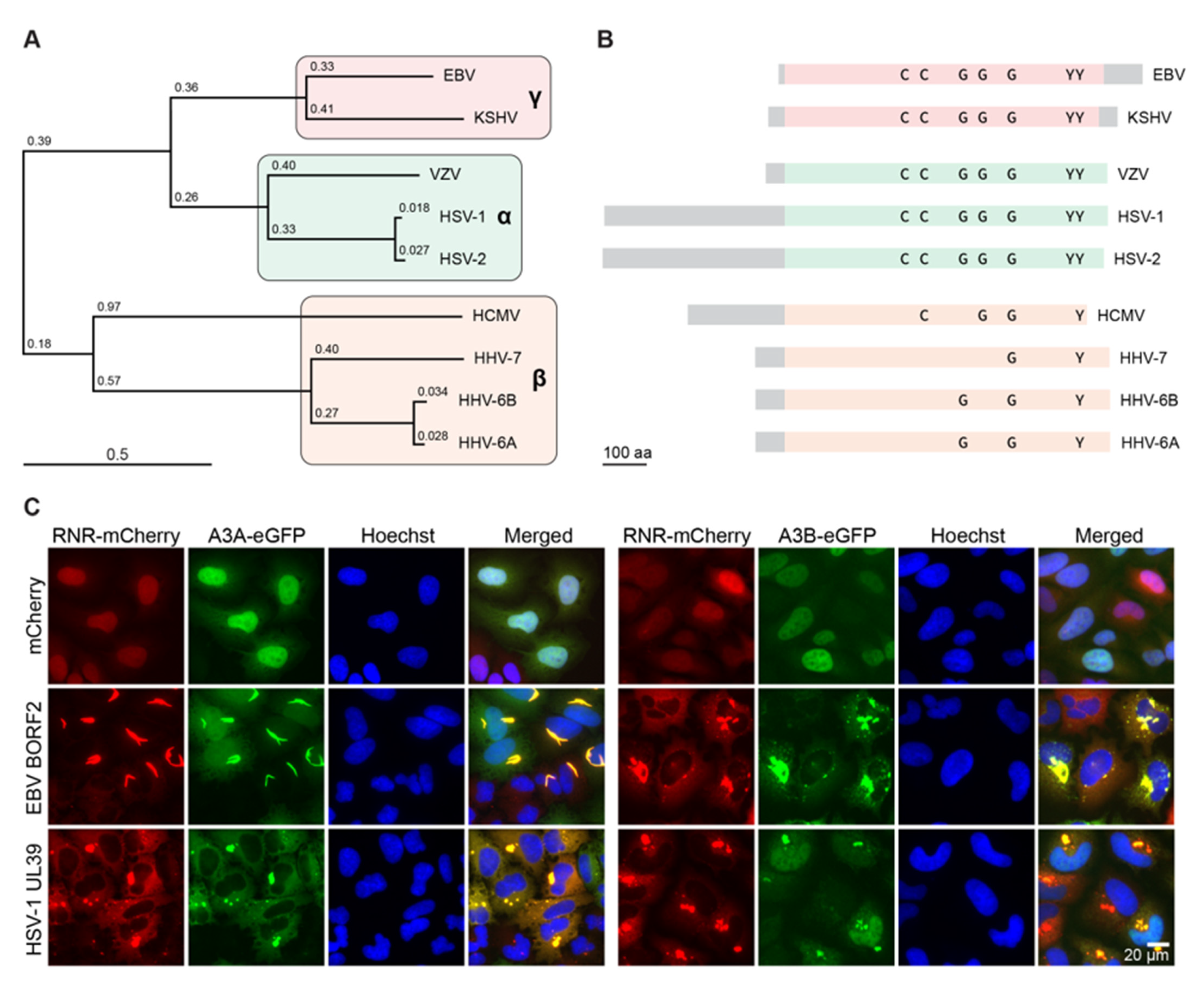
5. Evolutionary Perspectives
6. Clinical Implications
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Arvin, A.; Campadelli-Fiume, G.; Mocarski, E.; Moore, P.S.; Roizman, B.; Whitley, R.; Yamanishi, K. (Eds.) Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Grinde, B. Herpesviruses: Latency and reactivation - viral strategies and host response. J. Oral. Microbiol. 2013, 5, 22766. [Google Scholar] [CrossRef] [Green Version]
- Kieff, E.; Rickinson, A.B. Epstein-Barr virus and its replication. In Fields Virology; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2007; pp. 2603–2654. [Google Scholar]
- Nicoll, M.P.; Proenca, J.T.; Efstathiou, S. The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev. 2012, 36, 684–705. [Google Scholar] [CrossRef]
- Dyson, P.J.; Farrell, P.J. Chromatin structure of Epstein-Barr virus. J. Gen. Virol. 1985, 66, 1931–1940. [Google Scholar] [CrossRef]
- Bryant, H.; Farrell, P.J. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 2002, 76, 10290–10298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speck, S.H.; Chatila, T.; Flemington, E. Reactivation of Epstein-Barr virus: Regulation and function of the BZLF1 gene. Trends Microbiol. 1997, 5, 399–405. [Google Scholar] [CrossRef]
- Kang, M.S.; Kieff, E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoeger, T.; Adler, H. "Novel" triggers of herpesvirus reactivation and their potential health relevance. Front. Microbiol. 2018, 9, 3207. [Google Scholar] [CrossRef]
- Weller, S.K.; Coen, D.M. Herpes simplex viruses: Mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012, 4, a013011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mettenleiter, T.C. Herpesvirus assembly and egress. J. Virol. 2002, 76, 1537–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drake, J.W.; Hwang, C.B. On the mutation rate of herpes simplex virus type 1. Genetics 2005, 170, 969–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Hwang, Y.T.; Hwang, C.B. Mutation spectra of herpes simplex virus type 1 thymidine kinase mutants. J. Virol. 2002, 76, 5822–5828. [Google Scholar] [CrossRef] [Green Version]
- Renzette, N.; Pokalyuk, C.; Gibson, L.; Bhattacharjee, B.; Schleiss, M.R.; Hamprecht, K.; Yamamoto, A.Y.; Mussi-Pinhata, M.M.; Britt, W.J.; Jensen, J.D.; et al. Limits and patterns of cytomegalovirus genomic diversity in humans. Proc. Natl. Acad. Sci. USA 2015, 112, E4120–E4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjuan, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479-480, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chemudupati, M.; Kenney, A.D.; Bonifati, S.; Zani, A.; McMichael, T.M.; Wu, L.; Yount, J.S. From APOBEC to ZAP: Diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Radetskyy, R.; Daher, A.; Gatignol, A. ADAR1 and PKR, interferon stimulated genes with clashing effects on HIV-1 replication. Cytokine Growth Factor Rev. 2018, 40, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Locatelli, F. Adars: Allies or enemies? The importance of A-to-I RNA editing in human disease: From cancer to HIV-1. Biol. Rev. Camb. Philos. Soc. 2012, 87, 95–110. [Google Scholar] [CrossRef]
- Paladino, P.; Marcon, E.; Greenblatt, J.; Frappier, L. Identification of herpesvirus proteins that contribute to G1/S arrest. J. Virol. 2014, 88, 4480–4492. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.Z.; Yockteng-Melgar, J.; Jarvis, M.C.; Malik-Soni, N.; Borozan, I.; Carpenter, M.A.; McCann, J.L.; Ebrahimi, D.; Shaban, N.M.; Marcon, E.; et al. Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019, 4, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Refsland, E.W.; Stenglein, M.D.; Shindo, K.; Albin, J.S.; Brown, W.L.; Harris, R.S. Quantitative profiling of the full APOBEC3 mrna repertoire in lymphocytes and tissues: Implications for HIV-1 restriction. Nucleic Acids Res. 2010, 38, 4274–4284. [Google Scholar] [CrossRef] [Green Version]
- Brown, W.L.; Law, E.K.; Argyris, P.P.; Carpenter, M.A.; Levin-Klein, R.; Ranum, A.N.; Molan, A.M.; Forster, C.L.; Anderson, B.D.; Lackey, L.; et al. A rabbit monoclonal antibody against the antiviral and cancer genomic DNA mutating enzyme APOBEC3B. Antibodies 2019, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Lembo, D.; Brune, W. Tinkering with a viral ribonucleotide reductase. Trends Biochem. Sci. 2009, 34, 25–32. [Google Scholar] [CrossRef]
- Nordlund, P.; Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006, 75, 681–706. [Google Scholar] [CrossRef] [PubMed]
- Brignole, E.J.; Ando, N.; Zimanyi, C.M.; Drennan, C.L. The prototypic class Ia ribonucleotide reductase from Escherichia coli: Still surprising after all these years. Biochem. Soc. Trans. 2012, 40, 523–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Wu, S.Q.; Liang, Y.; Zhou, X.; Chen, W.; Li, L.; Wu, J.; Zhuang, Q.; Chen, C.; Li, J.; et al. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 2015, 17, 229–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langelier, Y.; Bergeron, S.; Chabaud, S.; Lippens, J.; Guilbault, C.; Sasseville, A.M.; Denis, S.; Mosser, D.D.; Massie, B. The R1 subunit of herpes simplex virus ribonucleotide reductase protects cells against apoptosis at, or upstream of, caspase-8 activation. J. Gen. Virol. 2002, 83, 2779–2789. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Bertrand, L.; Pearson, A.; Grandvaux, N.; Langelier, Y. The ribonucleotide reductase R1 subunits of herpes simplex virus 1 and 2 protect cells against poly(I · C)-induced apoptosis. J. Virol. 2011, 85, 8689–8701. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Omoto, S.; Harris, P.A.; Finger, J.N.; Bertin, J.; Gough, P.J.; Kaiser, W.J.; Mocarski, E.S. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 2015, 17, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufour, F.; Sasseville, A.M.; Chabaud, S.; Massie, B.; Siegel, R.M.; Langelier, Y. The ribonucleotide reductase R1 subunits of herpes simplex virus types 1 and 2 protect cells against TNFα- and FasL-induced apoptosis by interacting with caspase-8. Apoptosis 2011, 16, 256–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, D.; Pereira, E.F.; Gober, M.; Yarowsky, P.J.; Aurelian, L. The herpes simplex virus type 2 R1 protein kinase (ICP10 PK) blocks apoptosis in hippocampal neurons, involving activation of the MEK/MAPK survival pathway. J. Virol. 2002, 76, 1435–1449. [Google Scholar] [CrossRef] [Green Version]
- Brune, W.; Menard, C.; Heesemann, J.; Koszinowski, U.H. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 2001, 291, 303–305. [Google Scholar] [CrossRef]
- Mack, C.; Sickmann, A.; Lembo, D.; Brune, W. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc. Natl. Acad. Sci. USA 2008, 105, 3094–3099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, K.M.; Oh, S.E.; Kim, Y.E.; Han, T.H.; Ahn, J.H. Cooperative inhibition of RIP1-mediated NF-κB signaling by cytomegalovirus-encoded deubiquitinase and inactive homolog of cellular ribonucleotide reductase large subunit. PLoS Pathog. 2017, 13, e1006423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrone, M.; Percivalle, E.; Secchi, M.; Fiorina, L.; Pedrali-Noy, G.; Zoppe, M.; Baldanti, F.; Hahn, G.; Koszinowski, U.H.; Milanesi, G.; et al. The human cytomegalovirus UL45 gene product is a late, virion-associated protein and influences virus growth at low multiplicities of infection. J. Gen. Virol. 2003, 84, 3359–3370. [Google Scholar] [CrossRef] [PubMed]
- Silvas, T.V.; Schiffer, C.A. APOBEC3s: DNA-editing human cytidine deaminases. Protein Sci. 2019, 28, 1552–1566. [Google Scholar] [CrossRef]
- Green, A.M.; Weitzman, M.D. The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair (Amst) 2019, 83, 102700. [Google Scholar] [CrossRef] [PubMed]
- Janini, M.; Rogers, M.; Birx, D.R.; McCutchan, F.E. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 2001, 75, 7973–7986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, M.M.; Fahrny, A.; Jayaprakash, A.; Gers-Huber, G.; Dillon-White, M.; Audige, A.; Mulder, L.C.F.; Sachidanandam, R.; Speck, R.F.; Simon, V. Impact of suboptimal APOBEC3G neutralization on the emergence of HIV drug resistance in humanized mice. J. Virol. 2020, 94, 5. [Google Scholar] [CrossRef]
- Shi, K.; Carpenter, M.A.; Banerjee, S.; Shaban, N.M.; Kurahashi, K.; Salamango, D.J.; McCann, J.L.; Starrett, G.J.; Duffy, J.V.; Demir, O.; et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017, 24, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Kouno, T.; Silvas, T.V.; Hilbert, B.J.; Shandilya, S.M.D.; Bohn, M.F.; Kelch, B.A.; Royer, W.E.; Somasundaran, M.; Kurt Yilmaz, N.; Matsuo, H.; et al. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 2017, 8, 15024. [Google Scholar] [CrossRef] [Green Version]
- Wagner, J.R.; Demir, O.; Carpenter, M.A.; Aihara, H.; Harki, D.A.; Harris, R.S.; Amaro, R.E. Determinants of oligonucleotide selectivity of APOBEC3B. J. Chem. Inf. Model. 2018, 59, 2264–2273. [Google Scholar] [CrossRef]
- Goldstein, D.J.; Weller, S.K. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: Characterization of an ICP6 deletion mutant. Virology 1988, 166, 41–51. [Google Scholar] [CrossRef]
- Goldstein, D.J.; Weller, S.K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: Isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 1988, 62, 196–205. [Google Scholar] [CrossRef] [Green Version]
- Heineman, T.C.; Cohen, J.I. Deletion of the varicella-zoster virus large subunit of ribonucleotide reductase impairs growth of virus in vitro. J. Virol. 1994, 68, 3317–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.C.; Kulka, M.; Wymer, J.P.; Chung, T.D.; Aurelian, L. Expression of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) is required for virus growth and neoplastic transformation. J. Gen. Virol. 1992, 73 Pt 6, 1417–1428. [Google Scholar] [CrossRef]
- Mostafa, H.H.; Thompson, T.W.; Konen, A.J.; Haenchen, S.D.; Hilliard, J.G.; Macdonald, S.J.; Morrison, L.A.; Davido, D.J. Herpes simplex virus 1 mutant with point mutations in UL39 is impaired for acute viral replication in mice, establishment of latency, and explant-induced reactivation. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.Z.; Moraes, S.N.; Attarian, C.; Yockteng-Melgar, J.; Jarvis, M.C.; Biolatti, M.; Galitska, G.; Dell’Oste, V.; Frappier, L.; Bierle, C.J.; et al. A conserved mechanism of APOBEC3 relocalization by herpesviral ribonucleotide reductase large subunits. J. Virol. 2019, 93, 23. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Holland, T.C.; Bhagwat, A.S. Human herpes simplex virus-1 depletes APOBEC3A from nuclei. Virology 2019, 537, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Martinez, T.; Shapiro, M.; Bhaduri-McIntosh, S.; MacCarthy, T. Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses. Virus Evol. 2019, 5, vey040. [Google Scholar] [CrossRef] [PubMed]
- Minkah, N.; Chavez, K.; Shah, P.; Maccarthy, T.; Chen, H.; Landau, N.; Krug, L.T. Host restriction of murine gammaherpesvirus 68 replication by human APOBEC3 cytidine deaminases but not murine APOBEC3. Virology 2014, 454–455, 215–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulain, F.; Lejeune, N.; Willemart, K.; Gillet, N.A. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog. 2020, 16, e1008718. [Google Scholar] [CrossRef]
- Suspene, R.; Aynaud, M.M.; Koch, S.; Pasdeloup, D.; Labetoulle, M.; Gaertner, B.; Vartanian, J.P.; Meyerhans, A.; Wain-Hobson, S. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 2011, 85, 7594–7602. [Google Scholar] [CrossRef] [Green Version]
- Apseloff, G. Therapeutic uses of gallium nitrate: Past, present, and future. Am. J. Ther. 1999, 6, 327–339. [Google Scholar] [CrossRef]
- Lanzkron, S.; Strouse, J.J.; Wilson, R.; Beach, M.C.; Haywood, C.; Park, H.; Witkop, C.; Bass, E.B.; Segal, J.B. Systematic review: Hydroxyurea for the treatment of adults with sickle cell disease. Ann. Intern. Med. 2008, 148, 939–955. [Google Scholar] [CrossRef]
- Noble, S.; Goa, K.L. Gemcitabine. A review of its pharmacology and clinical potential in non-small cell lung cancer and pancreatic cancer. Drugs 1997, 54, 447–472. [Google Scholar] [CrossRef]
- Bernstein, L.R. Mechanisms of therapeutic activity for gallium. Pharmacol. Rev. 1998, 50, 665–682. [Google Scholar] [PubMed]
- Yarbro, J.W. Mechanism of action of hydroxyurea. Semin. Oncol. 1992, 19, 1–10. [Google Scholar]
- Plunkett, W.; Huang, P.; Xu, Y.Z.; Heinemann, V.; Grunewald, R.; Gandhi, V. Gemcitabine: Metabolism, mechanisms of action, and self-potentiation. Semin. Oncol. 1995, 22, 3–10. [Google Scholar]
- Kidd, J.M.; Newman, T.L.; Tuzun, E.; Kaul, R.; Eichler, E.E. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007, 3, e63. [Google Scholar] [CrossRef]
- Kimura, H.; Ito, Y.; Kawabe, S.; Gotoh, K.; Takahashi, Y.; Kojima, S.; Naoe, T.; Esaki, S.; Kikuta, A.; Sawada, A.; et al. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: Prospective analysis of 108 cases. Blood 2012, 119, 673–686. [Google Scholar] [CrossRef] [Green Version]
- Kimura, H.; Morishima, T.; Kanegane, H.; Ohga, S.; Hoshino, Y.; Maeda, A.; Imai, S.; Okano, M.; Morio, T.; Yokota, S.; et al. Prognostic factors for chronic active Epstein-Barr virus infection. J. Infect. Dis. 2003, 187, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.C.; Yuan, J.M. Epidemiology of nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 421–429. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer 2001, 94, 153–156. [Google Scholar] [CrossRef]
- Harabuchi, Y.; Takahara, M.; Kishibe, K.; Nagato, T.; Kumai, T. Extranodal natural killer/T-cell lymphoma, nasal type: Basic science and clinical progress. Front. Pediatr. 2019, 7, 141. [Google Scholar] [CrossRef]
- Aozasa, K.; Takakuwa, T.; Hongyo, T.; Yang, W.I. Nasal NK/T-cell lymphoma: Epidemiology and pathogenesis. Int. J. Hematol. 2008, 87, 110–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haverkos, B.M.; Pan, Z.; Gru, A.A.; Freud, A.G.; Rabinovitch, R.; Xu-Welliver, M.; Otto, B.; Barrionuevo, C.; Baiocchi, R.A.; Rochford, R.; et al. Extranodal NK/T cell lymphoma, nasal type (ENKTL-NT): An update on epidemiology, clinical presentation, and natural history in North American and European cases. Curr. Hematol. Malig. Rep. 2016, 11, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015, 5, 704–712. [Google Scholar] [CrossRef] [Green Version]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef]
- Venkatesan, S.; Rosenthal, R.; Kanu, N.; McGranahan, N.; Bartek, J.; Quezada, S.A.; Hare, J.; Harris, R.S.; Swanton, C. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018, 29, 563–572. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Schrijver, W.A.; Dalm, S.U.; de Weerd, V.; Moelans, C.B.; Ter Hoeve, N.; van Diest, P.J.; Martens, J.W.; van Deurzen, C.H. Progressive APOBEC3B mRNA expression in distant breast cancer metastases. PLOS ONE 2017, 12, e0171343. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Willis, S.; Burns, M.B.; Look, M.P.; Meijer-Van Gelder, M.E.; Schlicker, A.; Heideman, M.R.; Jacobs, H.; Wessels, L.; Leyland-Jones, B.; et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer 2014, 5, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Law, E.K.; Sieuwerts, A.M.; LaPara, K.; Leonard, B.; Starrett, G.J.; Molan, A.M.; Temiz, N.A.; Vogel, R.I.; Meijer-van Gelder, M.E.; Sweep, F.C.; et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci Adv. 2016, 2, e1601737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef]
- Burns, M.B.; Temiz, N.A.; Harris, R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013, 45, 977–983. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Buisson, R.; Langenbucher, A.; Bowen, D.; Kwan, E.E.; Benes, C.H.; Zou, L.; Lawrence, M.S. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 2019, 364, 6447. [Google Scholar] [CrossRef]
- Cortez, L.M.; Brown, A.L.; Dennis, M.A.; Collins, C.D.; Brown, A.J.; Mitchell, D.; Mertz, T.M.; Roberts, S.A. APOBEC3A is a prominent cytidine deaminase in breast cancer. PLoS Genet. 2019, 15, e1008545. [Google Scholar] [CrossRef] [Green Version]
- Maciejowski, J.; Chatzipli, A.; Dananberg, A.; Chu, K.; Toufektchan, E.; Klimczak, L.J.; Gordenin, D.A.; Campbell, P.J.; de Lange, T. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nat. Genet. 2020, 52, 884–890. [Google Scholar] [CrossRef]
- Law, E.K.; Levin-Klein, R.; Jarvis, M.C.; Kim, H.; Argyris, P.P.; Carpenter, M.A.; Starrett, G.J.; Temiz, N.A.; Larson, L.K.; Durfee, C.; et al. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo. J. Exp. Med. 2020, 217, 12. [Google Scholar] [CrossRef]
- Campadelli-Fiume, G.; De Giovanni, C.; Gatta, V.; Nanni, P.; Lollini, P.L.; Menotti, L. Rethinking herpes simplex virus: The way to oncolytic agents. Rev. Med. Virol. 2011, 21, 213–226. [Google Scholar] [CrossRef]
- Koch, M.S.; Lawler, S.E.; Chiocca, E.A. HSV-1 oncolytic viruses from bench to bedside: An overview of current clinical trials. Cancers 2020, 12, 3514. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S.L.; Emerman, M.; Malik, H.S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004, 2, E275. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Gifford, R.J.; Sato, K. Retroviruses drive the rapid evolution of mammalian APOBEC3 genes. Proc. Natl. Acad. Sci. USA 2020, 117, 610–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noguchi, S.; Arakawa, T.; Fukuda, S.; Furuno, M.; Hasegawa, A.; Hori, F.; Ishikawa-Kato, S.; Kaida, K.; Kaiho, A.; Kanamori-Katayama, M.; et al. FANTOM5 cage profiles of human and mouse samples. Sci. Data 2017, 4, 170112. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, A.Z.; Moraes, S.N.; Shaban, N.M.; Fanunza, E.; Bierle, C.J.; Southern, P.J.; Bresnahan, W.A.; Rice, S.A.; Harris, R.S. APOBECs and Herpesviruses. Viruses 2021, 13, 390. https://doi.org/10.3390/v13030390
Cheng AZ, Moraes SN, Shaban NM, Fanunza E, Bierle CJ, Southern PJ, Bresnahan WA, Rice SA, Harris RS. APOBECs and Herpesviruses. Viruses. 2021; 13(3):390. https://doi.org/10.3390/v13030390
Chicago/Turabian StyleCheng, Adam Z., Sofia N. Moraes, Nadine M. Shaban, Elisa Fanunza, Craig J. Bierle, Peter J. Southern, Wade A. Bresnahan, Stephen A. Rice, and Reuben S. Harris. 2021. "APOBECs and Herpesviruses" Viruses 13, no. 3: 390. https://doi.org/10.3390/v13030390
APA StyleCheng, A. Z., Moraes, S. N., Shaban, N. M., Fanunza, E., Bierle, C. J., Southern, P. J., Bresnahan, W. A., Rice, S. A., & Harris, R. S. (2021). APOBECs and Herpesviruses. Viruses, 13(3), 390. https://doi.org/10.3390/v13030390






