Adjusting the Cut-Off and Maximum Pool Size in RT-qPCR Pool Testing for SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Amplification Factor Estimation
2.3. Cut-Off Adaptation for Pooled Samples
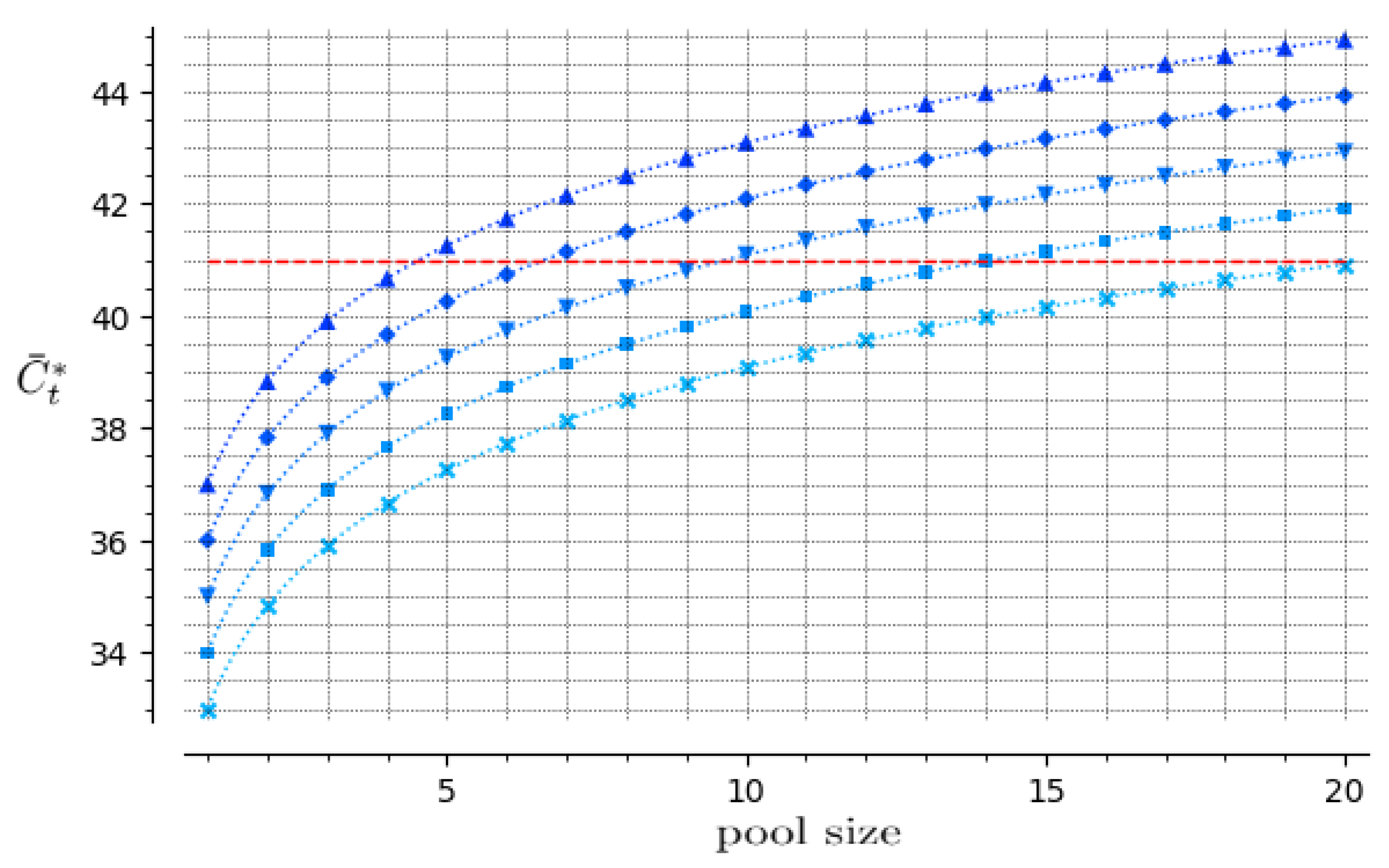
2.4. Pool Size Upper Bound
2.5. RT-qPCR Testing Framework
2.6. Calibration Experiments
2.7. Cut-Off Adjustment and Pool Size Choice
2.8. Pool Testing for Patients with Mild Symptoms
3. Results
4. Discussion
- Find an estimate of lower bound for the amplification factor value corresponding to the laboratory setting to be used, by a calibration assay that compares of individual and pooled samples, using Equation (3) and a lognormal parametric model of probability distribution.
- For each pool size, calculate the corresponding correction of cut-off value, according to Equation (4).
- Considering the equipment detection range and the desired target cut-off for individual testing, employ Equation (5) for finding the maximal admissible pool size.
- If the optimal pool size under the economic viewpoint, as stated in Reference [11], is greater than that maximal admissible pool size, adopt that maximal value; otherwise, employ the optimal size.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| # | Sample ID | Patient Initials | Age | Gender | Sample Collection Date | Sample Receipt Date | Pool # | Pool Size | Pool Plate ID | Pool Reaction Date | Pool Gene E Ct Value | Pool RNase P Ct Value | Pool RT-qPCR Result | Individual Plate ID | Individual Reaction Date | Individual Gene E Ct Value | Individual RNase P Ct Value | Individual Result |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P-A1 | AOD | 38 | Female | 10 September 2020 | 11 September 2020 | 1 | 12 | P332 | 11 September 2020 | 20.7 | 23.4 | Detected | P336 | 14 September 2020 | 37.4 | 22.0 | Undetected |
| 2 | P-A2 | FOH | 25 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 22.9 | Undetected | |||||||
| 3 | P-A3 | FDM | 67 | Female | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 22.4 | Undetected | |||||||
| 4 | P-A4 | SMR | 27 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 22.9 | Undetected | |||||||
| 5 | P-A5 | MSS | 47 | Female | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | 16.9 | 22.3 | Detected | |||||||
| 6 | P-A6 | FCN | 38 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 23.0 | Undetected | |||||||
| 7 | P-A7 | MMG | 37 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 21.2 | Undetected | |||||||
| 8 | P-A8 | ISN | 30 | Female | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 21.2 | Undetected | |||||||
| 9 | P-A10 | WASM | 32 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 26.8 | Undetected | |||||||
| 10 | P-A11 | ACRA | 56 | Female | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 22.4 | Undetected | |||||||
| 11 | P-A12 | FFD | 62 | Female | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 24.6 | Undetected | |||||||
| 12 | P-A13 | JSF | 32 | Male | 10 September 2020 | 11 September 2020 | P336 | 14 September 2020 | Undetermined | 24.3 | Undetected | |||||||
| 77 | P-A105 | VFC | 48 | Female | 22 September 2020 | 23 September 2020 | 8 | 8 | P354 | 23 September 2020 | 23.1 | 25.5 | Detected | P362 | 28 September 2020 | Undetermined | 26.6 | Undetected |
| 78 | P-A106 | PMAA | 42 | Female | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 25.8 | Undetected | |||||||
| 79 | P-A107 | FSC | 31 | Female | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 26.7 | Undetected | |||||||
| 80 | P-A108 | EPAF | 34 | Male | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 25.8 | Undetected | |||||||
| 81 | P-A109 | LPSJ | 22 | Male | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 23.1 | Undetected | |||||||
| 82 | P-A110 | CORA | 19 | Female | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 26.5 | Undetected | |||||||
| 83 | P-A111 | RSP | 20 | Female | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | Undetermined | 26.5 | Undetected | |||||||
| 84 | P-A112 | AGPJM | 39 | Male | 22 September 2020 | 23 September 2020 | P355 | 24 September 2020 | 18.1 | 25.5 | Detected | |||||||
| 109 | P-A146 | SCJ | 34 | Male | 25 September 2020 | 28 September 2020 | 14 | 3 | P362 | 28 September 2020 | 28.9 | 25.2 | Detected | P365 | 29 September 2020 | Undetermined | 25.0 | Undetected |
| 110 | P-A147 | AOMC | 34 | Male | 25 September 2020 | 28 September 2020 | P365 | 29 September 2020 | Undetermined | 25.5 | Undetected | |||||||
| 111 | P-A148 | AROA | 25 | Female | 25 September 2020 | 28 September 2020 | P365 | 29 September 2020 | 27.5 | 25.3 | Detected | |||||||
| 112 | P-A149 | DQM | 36 | Male | 25 September 2020 | 28 September 2020 | 15 | 3 | P362 | 28 September 2020 | 18.2 | 24.6 | Detected | P365 | 29 September 2020 | Undetermined | 24.5 | Undetected |
| 113 | P-A150 | SPK | 25 | Female | 25 September 2020 | 28 September 2020 | P365 | 29 September 2020 | 16.6 | 25.0 | Detected | |||||||
| 114 | P-A151 | FMM | 65 | Female | 25 September 2020 | 28 September 2020 | P365 | 29 September 2020 | Undetermined | 24.9 | Undetected | |||||||
| 118 | P-A191 | DCD | 61 | Male | 30 September 2020 | 1 October 2020 | 29 | 3 | P369 | 1 October 2020 | 25.0 | 25.3 | Detected | P372 | 2 October 2020 | Undetermined | 28.1 | Undetected |
| 119 | P-A192 | RBR | 61 | Female | 30 September 2020 | 1 October 2020 | P372 | 2 October 2020 | Undetermined | 25.8 | Undetected | |||||||
| 120 | P-A193 | NAA | 38 | Female | 30 September 2020 | 1 October 2020 | P372 | 2 October 2020 | 22.6 | 25.3 | Detected | |||||||
| 127 | P-A200 | CSC | 45 | Female | 2 October 2020 | 5 October 2020 | 32 | 3 | P374 | 5 October 2020 | 33.5 | 28.4 | Detected | P375 | 6 October 2020 | Undetermined | 28.5 | Undetected |
| 128 | P-A201 | JF | 41 | Male | 2 October 2020 | 5 October 2020 | P375 | 6 October 2020 | 30.8 | 29.3 | Detected | |||||||
| 129 | P-A202 | FFI | 73 | Male | 2 October 2020 | 5 October 2020 | P375 | 6 October 2020 | Undetermined | 28.0 | Undetected | |||||||
| 130 | P-A203 | SOE | 24 | Female | 2 October 2020 | 5 October 2020 | 33 | 3 | P374 | 5 October 2020 | 24.9 | 29.2 | Detected | P375 | 6 October 2020 | Undetermined | 28.9 | Undetected |
| 131 | P-A204 | FSA | 44 | Female | 2 October 2020 | 5 October 2020 | P375 | 6 October 2020 | Undetermined | 28.4 | Undetected | |||||||
| 132 | P-A205 | MSN | 44 | Male | 2 October 2020 | 5 October 2020 | P375 | 6 October 2020 | 22.5 | 28.4 | Detected | |||||||
| 133 | P-A209 | KTSP | 32 | Female | 2 October 2020 | 5 October 2020 | 35 | 8 | P375 | 6 October 2020 | 24.3 | 28.3 | Detected | P378 | 7 October 2020 | Undetermined | 30.3 | Undetected |
| 134 | P-A210 | SR | 53 | Male | 2 October 2020 | 5 October 2020 | P378 | 7 October 2020 | Undetermined | 27.9 | Undetected | |||||||
| 135 | P-A211 | FCGBVC | 26 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 28.0 | Undetected | |||||||
| 136 | P-A212 | PASA | 36 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | 21.0 | 29.1 | Detected | |||||||
| 137 | P-A213 | DSJ | 35 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 30.0 | Undetected | |||||||
| 138 | P-A214 | FGVG | 26 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 30.8 | Undetected | |||||||
| 139 | P-A215 | GSMM | 29 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 28.5 | Undetected | |||||||
| 140 | P-A216 | RSM | 45 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 28.3 | Undetected | |||||||
| 141 | P-A217 | GCJ | 37 | Male | 5 October 2020 | 6 October 2020 | 36 | 8 | P375 | 6 October 2020 | 40.5 | 27.5 | Detected | P378 | 7 October 2020 | Undetermined | 30.2 | Undetected |
| 142 | P-A218 | AFS | 35 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 29.6 | Undetected | |||||||
| 143 | P-A219 | LAL | 63 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 27.8 | Undetected | |||||||
| 144 | P-A220 | PESM | 27 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 27.5 | Undetected | |||||||
| 145 | P-A221 | CBSL | 29 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 26.7 | Undetected | |||||||
| 146 | P-A222 | VASM | 20 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 27.0 | Undetected | |||||||
| 147 | P-A223 | ARM | 31 | Male | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | 35.4 | 27.2 | Detected | |||||||
| 148 | P-A224 | SMP | 29 | Female | 5 October 2020 | 6 October 2020 | P378 | 7 October 2020 | Undetermined | 25.6 | Undetected | |||||||
| 149 | P-A225 | SPS | 22 | Female | 5 October 2020 | 6 October 2020 | 37 | 8 | P378 | 7 October 2020 | 28.1 | 27.7 | Detected | P379 | 8 October 2020 | 38.7 | 28.6 | Undetected |
| 150 | P-A226 | ASC | 46 | Female | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 27.7 | Undetected | |||||||
| 151 | P-A227 | SMW | 20 | Male | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 28.7 | Undetected | |||||||
| 152 | P-A228 | SCW | 28 | Male | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 28.5 | Undetected | |||||||
| 153 | P-A229 | FSK | 31 | Female | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | 26.4 | 28.9 | Detected | |||||||
| 154 | P-A230 | ROSW | 46 | Male | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 28.9 | Undetected | |||||||
| 155 | P-A231 | PSG | 22 | Male | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 29.5 | Undetected | |||||||
| 156 | P-A232 | JSCM | 45 | Female | 6 October 2020 | 7 October 2020 | P379 | 8 October 2020 | Undetermined | 27.0 | Undetected | |||||||
| 161 | P-A261 | OCH | 37 | Male | 9 October 2020 | 13 October 2020 | 43 | 4 | P384 | 13 October 2020 | 22.1 | 28.3 | Detected | P385 | 14 October 2020 | 18.0 | 27.9 | Detected |
| 162 | P-A262 | MCF | 36 | Female | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 26.5 | Undetected | |||||||
| 163 | P-A263 | GRE | 41 | Male | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 28.9 | Undetected | |||||||
| 164 | P-A264 | NTC | 26 | Female | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 29.7 | Undetected | |||||||
| 165 | P-A265 | ENL | 34 | Male | 9 October 2020 | 13 October 2020 | 44 | 4 | P384 | 13 October 2020 | 22.0 | 27.9 | Detected | P385 | 14 October 2020 | 18.5 | 25.4 | Detected |
| 166 | P-A266 | CPD | 42 | Female | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 27.0 | Undetected | |||||||
| 167 | P-A267 | MOG | 70 | Male | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 30.0 | Undetected | |||||||
| 168 | P-A268 | ABON | 31 | Male | 9 October 2020 | 13 October 2020 | P385 | 14 October 2020 | Undetermined | 29.9 | Undetected | |||||||
| 169 | P-A674 | RLA | 42 | Female | 30 November 2020 | 1 December 2020 | 177 | 3 | P468 | 1 December 2020 | 37.5 | 26.6 | Detected | P470 | 2 December 2020 | Undetermined | 24.6 | Undetected |
| 170 | P-A675 | PBM | 34 | Male | 30 November 2020 | 1 December 2020 | P375 | 2 December 2020 | Undetermined | 27.6 | Undetected | |||||||
| 171 | P-A676 | PHS | 35 | Male | 30 November 2020 | 1 December 2020 | P375 | 2 December 2020 | 34.8 | 26.9 | Detected |
References
- Cherif, A.; Grobe, N.; Wang, X.; Kotanko, P. Simulation of pool testing to identify patients with Coronavirus Disease 2019 under conditions of limited test availability. JAMA Netw. Open 2020, 3, e2013075. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. The mathematical strategy that could transform coronavirus testing. Nature 2020, 583, 504–505. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Lee, J.; Sung, H.; Lee, H.; Han, M.G.; Yoo, C.K.; Lee, S.W.; Hong, K.H. Pooling upper respiratory specimens for rapid mass screening of COVID-19 by real-time RT-PCR. Emerg Infect. Dis. 2020, 26, 2469–2472. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.; Albert, E.; Navarro, D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J. Med. Virol. 2020, 92, 2306–2307. [Google Scholar] [CrossRef] [PubMed]
- Yelin, I.; Aharony, N.; Tamar, E.S.; Argoetti, A.; Messer, E.; Berenbaum, D.; Shafran, E.; Kuzli, A.; Gandali, N.; Shkedi, O.; et al. Evaluation of COVID-19 RT-qPCR test in multi sample pools. Clin. Infect. Dis. 2020, 71, 2073–2078. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.E.; Lee, F.M. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Screening with Specimen Pools: Time to Swim, or Too Deep for Comfort? Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mutesa, L.; Ndishimye, P.; Butera, Y.; Souopgui, J.; Uwineza, A.; Rutayisire, R.; Ndoricimpaye, E.L.; Musoni, E.; Rujeni, N.; Nyatanyi, T.; et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature 2020, 589, 276–280. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleiker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 2020, 25, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, C.; Kanjilal, S.; Baker, M.; Klompas, M. Duration of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectivity: When Is It Safe to Discontinue Isolation? Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, R.; Aherfi, S.; Wurtz, N.; Grimaldier, C.; Hoang, V.T.; Colson, P.; Raoult, D.; Scola, B.L. Correlation between 3790 quantitative polymerase chain reaction–positives samples and positive cell cultures, including 1941 severe acute respiratory syndrome coronavirus 2 isolates. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Dorfman, R. The detection of defective members of large populations. Ann. Math. Stat. 1943, 14, 436–440. [Google Scholar] [CrossRef]
| Pool Size | Value of Individual Sample 1 | Values Obtained with Each Pool | Empirical Increase of | Recommended Increase in Cut-Off Value: |
|---|---|---|---|---|
| N | ||||
| 12 | 16.9 | 20.7 | 3.8 | 6.6 |
| 8 | 18.1 | 23.1 | 5.0 | 5.5 |
| 8 | 21.0 | 24.3 | 3.3 | 5.5 |
| 8 | 35.4 | 40.5 | 5.1 | 5.5 |
| 8 | 26.4 | 28.1 | 1.7 | 5.5 |
| 3 | 34.8 | 37.5 | 2.7 | 2.9 |
| 3 | 27.5 | 28.9 | 1.4 | 2.9 |
| 3 | 16.6 | 18.2 | 1.6 | 2.9 |
| 3 | 30.8 | 33.5 | 2.7 | 2.9 |
| 3 | 22.6 | 25.0 | 2.4 | 2.9 |
| 3 | 22.5 | 24.9 | 2.4 | 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, M.S.; Sato, H.I.; Rocha, R.P.; Carvalho, A.F.; Guimarães, N.S.; Machado, E.L.; Alves, C.R.L.; Teixeira, S.M.R.; Takahashi, R.H.C.; Tupinambás, U.; et al. Adjusting the Cut-Off and Maximum Pool Size in RT-qPCR Pool Testing for SARS-CoV-2. Viruses 2021, 13, 557. https://doi.org/10.3390/v13040557
Costa MS, Sato HI, Rocha RP, Carvalho AF, Guimarães NS, Machado EL, Alves CRL, Teixeira SMR, Takahashi RHC, Tupinambás U, et al. Adjusting the Cut-Off and Maximum Pool Size in RT-qPCR Pool Testing for SARS-CoV-2. Viruses. 2021; 13(4):557. https://doi.org/10.3390/v13040557
Chicago/Turabian StyleCosta, Murilo S., Hugo I. Sato, Raissa P. Rocha, Alex F. Carvalho, Nathalia S. Guimarães, Elaine L. Machado, Claudia R. L. Alves, Santuza M. R. Teixeira, Ricardo H. C. Takahashi, Unaí Tupinambás, and et al. 2021. "Adjusting the Cut-Off and Maximum Pool Size in RT-qPCR Pool Testing for SARS-CoV-2" Viruses 13, no. 4: 557. https://doi.org/10.3390/v13040557
APA StyleCosta, M. S., Sato, H. I., Rocha, R. P., Carvalho, A. F., Guimarães, N. S., Machado, E. L., Alves, C. R. L., Teixeira, S. M. R., Takahashi, R. H. C., Tupinambás, U., & da Fonseca, F. G. (2021). Adjusting the Cut-Off and Maximum Pool Size in RT-qPCR Pool Testing for SARS-CoV-2. Viruses, 13(4), 557. https://doi.org/10.3390/v13040557








