Phylogenetic Clustering among Asylum Seekers with New HIV-1 Diagnoses in Montreal, QC, Canada
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Setting, and Participants
2.2. Data Collection and Variables of Interest
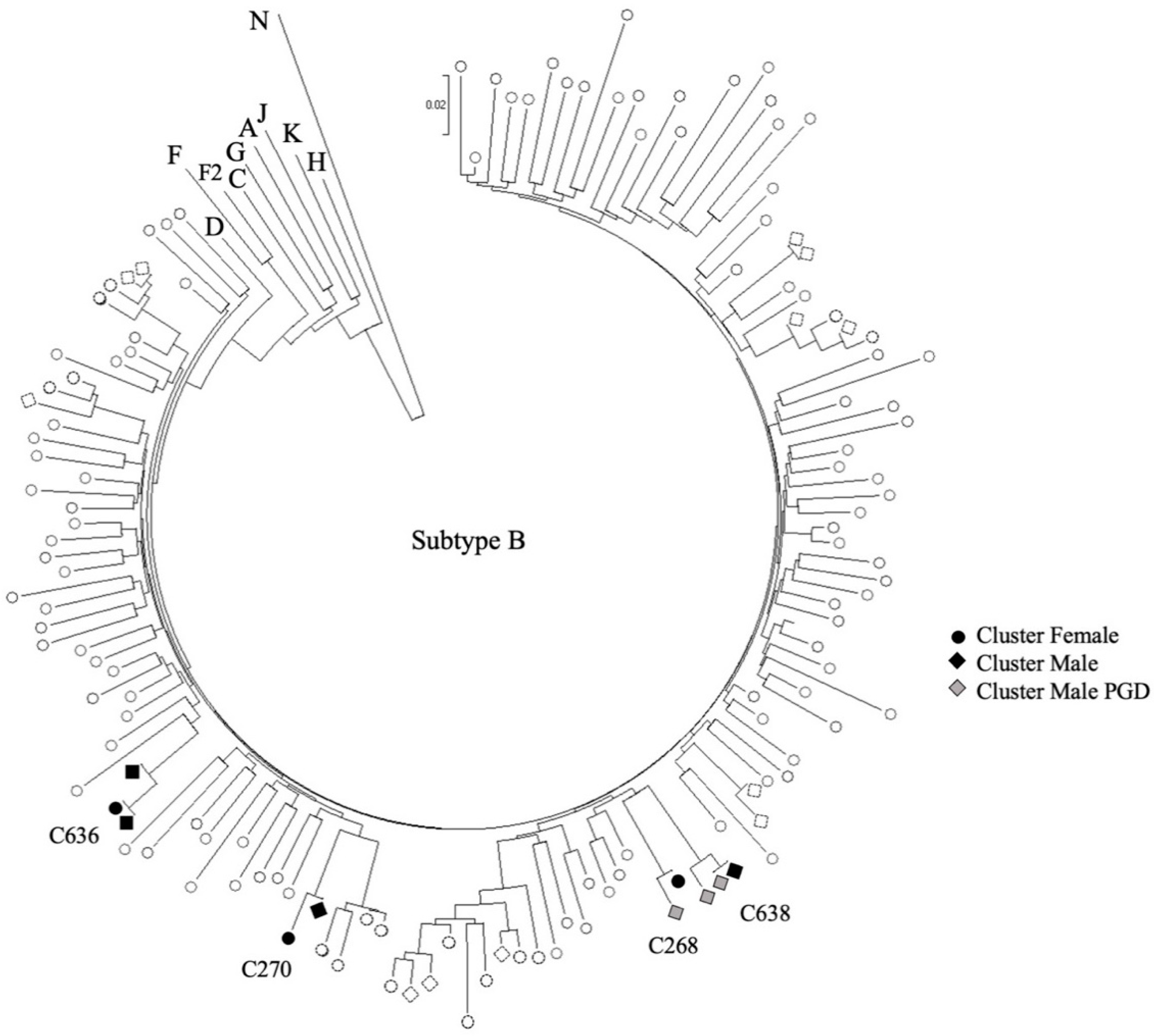
2.3. Phylogenetic Analysis
2.4. Statistical Analyses
3. Results
3.1. Patient Characteristics
3.2. Transmission Clusters
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alvarez-del Arco, D.; Monge, S.; Azcoaga, A.; Rio, I.; Hernando, V.; Gonzalez, C.; Alejos, B.; Caro, A.M.; Perez-Cachafeiro, S.; Ramirez-Rubio, O.; et al. HIV testing and counselling for migrant populations living in high-income countries: A systematic review. Eur. J. Public Health 2013, 23, 1039–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakoya, I.; Álvarez-del Arco, D.; Woode-Owusu, M.; Monge, S.; Rivero-Montesdeoca, Y.; Delpech, V.; Rice, B.; Noori, T.; Pharris, A.; Amato-Gauci, A.J.; et al. A systematic review of post-migration acquisition of HIV among migrants from countries with generalised HIV epidemics living in Europe: Mplications for effectively managing HIV prevention programmes and policy. BMC Public Health 2015, 15, 561. [Google Scholar] [CrossRef] [Green Version]
- Gunaratnam, P.; Heywood, A.E.; McGregor, S.; Jamil, M.S.; McManus, H.; Mao, L.; Lobo, R.; Brown, G.; Hellard, M.; Marukutira, T.; et al. HIV diagnoses in migrant populations in Australia—A changing epidemiology. PLoS ONE 2019, 14, e0212268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haddad, N.; Robert, A.; Weeks, A.; Popovic, N.; Siu, W.; Archibald, C. HIV in Canada-Surveillance Report, 2018. Can. Commun. Dis. Rep. 2019, 45, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.; Li, J.S.; Totten, S.; McGuire, M. HIV in Canada-Surveillance Report, 2017. Can. Commun. Dis. Rep. 2018, 44, 348–356. [Google Scholar] [CrossRef]
- Mercure, S.-A.; (Direction Régionale de Santé Publique, CIUSSS Centre-Sud-de-l’Île-de-Montréal, Montréal, QC, Canada). Personal communication, 2020.
- Kronfli, N.; Linthwaite, B.; Sheehan, N.; Cox, J.; Hardy, I.; Lebouché, B.; de Pokomandy, A.; Frenette, C.; Roger, M.; Klein, M.B. Delayed linkage to HIV care among asylum seekers in Quebec, Canada. BMC Public Health 2019, 19, 1683. [Google Scholar] [CrossRef]
- Alvarez-Del Arco, D.; Fakoya, I.; Thomadakis, C.; Pantazis, N.; Touloumi, G.; Gennotte, A.F.; Zuure, F.; Barros, H.; Staehelin, C.; Göpel, S.; et al. High levels of postmigration HIV acquisition within nine European countries. AIDS 2017, 31, 1979–1988. [Google Scholar] [CrossRef] [Green Version]
- Pantazis, N.; Thomadakis, C.; Del Amo, J.; Alvarez-Del Arco, D.; Burns, F.M.; Fakoya, I.; Touloumi, G. Determining the likely place of HIV acquisition for migrants in Europe combining subject-specific information and biomarkers data. Stat. Methods Med. Res. 2019, 28, 1979–1997. [Google Scholar] [CrossRef] [Green Version]
- Sacks-Davis, R.; Chibo, D.; Peach, E.; Aleksic, E.; Crowe, S.M.; El Hayek, C.; Marukutira, T.; Higgins, N.; Stoove, M.; Hellard, M. Phylogenetic clustering networks among heterosexual migrants with new HIV diagnoses post-migration in Australia. PLoS ONE 2020, 15, e0237469. [Google Scholar] [CrossRef]
- Op de Coul, E.L.M.; van Sighem, A.; Brinkman, K.; van Benthem, B.H.; van der Ende, M.E.; Geerlings, S.; Reiss, P.; Cohort, A.N.O.H. Factors associated with presenting late or with advanced HIV disease in the Netherlands, 1996–2014: Results from a national observational cohort. BMJ Open 2016, 6, e009688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J.; Cunningham, C.O.; Hanna, D.B. HIV outcomes among migrants from low-income and middle-income countries living in high-income countries: A review of recent evidence. Curr. Opin. Infect. Dis. 2018, 31, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Blais, M.; Fernet M Fau-Proulx-Boucher, K.; Proulx-Boucher, K.; Fau-Lebouché, B.; Lebouché, B.; Fau-Rodrigue, C.; Rodrigue, C.; Fau-Lapointe, N.; Lapointe, N.; Fau-Otis, J.; et al. Barriers to health-care and psychological distress among mothers living with HIV in Quebec (Canada). AIDS Care 2015, 27, 731–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nkulu Kalengayi, F.K.; Hurtig, A.-K.; Ahlm, C.; Krantz, I. Fear of deportation may limit legal immigrants’ access to HIV/AIDS-related care: A survey of Swedish language school students in Northern Sweden. J. Immigr. Minor. Health 2012, 14, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Brenner, B.G.; Roger, M.; Routy, J.-P.; Moisi, D.; Ntemgwa, M.; Matte, C.; Baril, J.-G.; Thomas, R.; Rouleau, D.; Bruneau, J.; et al. High Rates of Forward Transmission Events after Acute/Early HIV-1 Infection. J. Infect. Dis. 2007, 195, 951–959. [Google Scholar] [CrossRef]
- Villandré, L.; Labbe, A.; Brenner, B.; Ibanescu, R.-I.; Roger, M.; Stephens, D.A. Assessing the role of transmission chains in the spread of HIV-1 among men who have sex with men in Quebec, Canada. PLoS ONE 2019, 14, e0213366. [Google Scholar]
- Xiridou, M.; van Veen, M.; Coutinho, R.; Prins, M. Can migrants from high-endemic countries cause new HIV outbreaks among heterosexuals in low-endemic countries? AIDS 2010, 24, 13. [Google Scholar] [CrossRef]
- HIV Drug Resistance Database. Available online: https://hivdb.stanford.edu/ (accessed on 24 February 2021).
- Fonner, V.A.; Sands, A.; Figueroa, C.; Baggaley, R.; Quinn, C.; Jamil, M.S.; Johnson, C. Country adherence to WHO recommendations to improve the quality of HIV diagnosis: A global policy review. BMJ Glob. Health 2020, 5, e001939. [Google Scholar] [CrossRef] [PubMed]
- Staveteig, S.; Croft, T.N.; Kampa, K.T.; Head, S.K. Reaching the ‘first 90′: Gaps in coverage of HIV testing among people living with HIV in 16 African countries. PLoS ONE 2017, 12, e0186316. [Google Scholar] [CrossRef] [Green Version]
- UNAIDS. 90-90-90 An ambitious treatment target to help end the AIDS epidemic. In Joint United Nations Programme on HIV/AIDS; UNAIDS: Geneva, Switzerland, 2014. [Google Scholar]
- CDC HIV Cluster and Outbreak Detection and Response. Available online: https://www.cdc.gov/hiv/programresources/guidance/cluster-outbreak/index.html (accessed on 24 February 2021).
- McLaughlin, A.; Sereda, P.; Oliveira, N.; Barrios, R.; Brumme, C.J.; Brumme, Z.L.; Montaner, J.S.G.; Joy, J.B. Detection of HIV transmission hotspots in British Columbia, Canada: A novel framework for the prioritization and allocation of treatment and prevention resources. EBioMedicine 2019, 48, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, A.F.Y.; Gustafson, R.; Daly, P.; Zerr, L.; Demlow, S.E.; Wong, J.; Woods, C.K.; Hogg, R.S.; Krajden, M.; Moore, D.; et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: An implementation case study. Lancet HIV 2016, 3, e231–e238. [Google Scholar] [CrossRef] [Green Version]
- Wertheim, J.O.; Chato, C.; Poon, A.F.Y. Comparative analysis of HIV sequences in real time for public health. Cur. Opin. HIV AIDS 2019, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Gorbach, P.M.; Javanbakht, M.; Bolan, R.K. Behavior change following HIV diagnosis: Findings from a Cohort of Los Angeles MSM. AIDS Care 2018, 30, 300–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, E.; Shao, W.; Bontell, I.; Cham, F.; Cuong, D.D.; Wondwossen, A.; Morris, L.; Hunt, G.; Sönnerborg, A.; Bertagnolio, S.; et al. Evaluation of sequence ambiguities of the HIV-1 pol gene as a method to identify recent HIV-1 infection in transmitted drug resistance surveys. Infect. Genet. Evol. 2013, 18, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, B.G.; Ibanescu, R.I.; Hardy, I.; Stephens, D.; Otis, J.; Moodie, E.; Grossman, Z.; Vandamme, A.M.; Roger, M.; Wainberg, M.A. Large cluster outbreaks sustain the HIV epidemic among MSM in Quebec. AIDS 2017, 31, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Aris-Brosou, S.; Joanisse, I.; Merks, H.; Vallée, D.; Caminiti, K.; Rekart, M.; Krajden, M.; Cook, D.; Kim, J.; et al. Genetic Diversity as a Marker for Timing Infection in HIV-Infected Patients: Evaluation of a 6-Month Window and Comparison With BED. J. Infect. Dis. 2012, 206, 756–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Overall (n = 105) | MUHC (n = 83) | JGH (n = 22) | |
|---|---|---|---|
| Age (median (IQR)) | 41 [35;47] | 40 [35;46] | 44 [39;51] |
| Sex | |||
| Female | 51 (49%) | 46 (55%) | 5 (23%) |
| Sexual orientation | |||
| Heterosexual | 91 (87%) | 70 (84%) | 21 (95%) |
| LGBTQ | 14 (13%) | 13 (16%) | 1 (5%) |
| Birth country | |||
| Africa | |||
| Burundi | 4 (4%) | 2 (2%) | 2 (9%) |
| DRC | 6 (6%) | 5 (6%) | 1 (4%) |
| Nigeria | 18 (17%) | 18 (22%) | 0 (0%) |
| Other | 23 (22%) | 20 (24%) | 3 (14%) |
| Americas | |||
| Haiti | 48 (45%) | 36 (44%) | 12 (55%) |
| Other | 6 (6%) | 2 (2%) | 4 (18%) |
| HIV subtype | |||
| AG | 19 (18%) | 17 (21%) | 2 (9%) |
| B | 52 (50%) | 37 (45%) | 15 (68%) |
| C | 14 (13%) | 12 (14%) | 2 (9%) |
| G | 6 (6%) | 6 (7%) | 0 (0%) |
| Other | 14 (13%) | 11 (13%) | 3 (14%) |
| CD4 at diagnosis, cells/μL (median; range; (IQR)) | 303; 7–1567; [181;416] | 308; 7–811; [194;392] | 221; 29–1567; [159;474] |
| CD4 < 200 | 31 (30%) | 21 (25%) | 10 (45%) |
| CD4 200–350 | 34 (32%) | 29 (35%) | 5 (23%) |
| CD4 > 350 | 40 (38%) | 33 (40%) | 7 (32%) |
| VL at diagnosis, copies/mL (median; range; (IQR)) | 32,434; <20–1,348,292; [7429;80,017] | 32,434; <20–1,348,292; [6962;95,958] | 33,913; 89–347,068; [11,244;73,649] |
| VL 0–99,999 | 84 (80%) | 65 (78%) | 19 (86%) |
| VL 100,000–499,999 | 16 (15%) | 13 (16%) | 3 (14%) |
| VL ≥ 500,000 | 5 (5%) | 5 (6%) | 0 (0%) |
| Cluster Identification | HIV Subtype | Birth Country | Sex | Self Reported Sexual Orientation | Date of Arrival in Canada | Date of Sampling | CD4 at Genotype (cells/μL) | VL at Genotype (copies/mL) | Resistance Mutations |
|---|---|---|---|---|---|---|---|---|---|
| Clusters with Study Participants Only | |||||||||
| C636.01 | B | Haiti | M | HET | 08/2017 | 10/2017 | 167 | 32,908 | K103N |
| C636.02 | Haiti | M | HET | 10/2017 | 11/2017 | 347 | 99,274 | K103N | |
| C636.03 | Haiti | F | HET | 09/2017 | 12/2017 | 687 | 19,896 | K103N | |
| CT270.01 | B | Haiti | M | HET | 09/2017 | 11/2017 | 209 | 2549 | WT |
| CT270.02 | Haiti | F | HET | 09/2017 | 11/2017 | 191 | 68,743 | WT | |
| CAG41.01 | AG | Nigeria | M | HET | 12/2017 | 04/2018 | 333 | 181,853 | WT |
| CAG41.02 | Nigeria | F | HET | 12/2017 | 04/2018 | 316 | 11,524 | WT | |
| G5.01 | G | Nigeria | M | HET | 12/2017 | 02/2018 | 11 | 112,947 | M184V, T215F, G190A |
| G5.02 | Nigeria | F | HET | 12/2017 | 02/2018 | 531 | 40,988 | WT | |
| Clusters with Study Participants and Individuals from the Provincial Genotyping Database | |||||||||
| CT268.01 | B | N/A | M | N/A | N/A | 08/2017 | N/A | 1628 | K103KN |
| CT268.02 | Haiti | F | HET | 04/2018 | 04/2018 | 382 | 5487 | K103KN | |
| C569.01 | B | N/A | M | N/A | N/A | 08/2009 | N/A | 12,158 | WT |
| C569.02 | N/A | F | N/A | N/A | 03/2016 | N/A | 959 | WT | |
| C569.03 | Saint Vincent | F | HET | 06/2018 | 06/2018 | 122 | 15,752 | WT | |
| C638.01 | B | Haiti | M | MSM | 08/2017 | 11/2017 | 71 | 1,154,977 | WT |
| C638.02 | N/A | M | N/A | N/A | 02/2018 | N/A | 225,274 | WT | |
| C638.03 | N/A | M | N/A | N/A | 02/2018 | N/A | 238,744 | WT | |
| C645.01 | B | N/A | M | N/A | N/A | 02/2011 | N/A | 385,982 | WT |
| C645.02 | Algeria | M | MSM | 11/2017 | 06/2018 | 279 | 460,002 | WT | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.; Brenner, B.; Ibanescu, R.-I.; Cox, J.; Weiss, K.; Klein, M.B.; Hardy, I.; Narasiah, L.; Roger, M.; Kronfli, N. Phylogenetic Clustering among Asylum Seekers with New HIV-1 Diagnoses in Montreal, QC, Canada. Viruses 2021, 13, 601. https://doi.org/10.3390/v13040601
Park H, Brenner B, Ibanescu R-I, Cox J, Weiss K, Klein MB, Hardy I, Narasiah L, Roger M, Kronfli N. Phylogenetic Clustering among Asylum Seekers with New HIV-1 Diagnoses in Montreal, QC, Canada. Viruses. 2021; 13(4):601. https://doi.org/10.3390/v13040601
Chicago/Turabian StylePark, Hyejin, Bluma Brenner, Ruxandra-Ilinca Ibanescu, Joseph Cox, Karl Weiss, Marina B. Klein, Isabelle Hardy, Lavanya Narasiah, Michel Roger, and Nadine Kronfli. 2021. "Phylogenetic Clustering among Asylum Seekers with New HIV-1 Diagnoses in Montreal, QC, Canada" Viruses 13, no. 4: 601. https://doi.org/10.3390/v13040601






