Structural Characterization of a Minimal Antibody against Human APOBEC3B
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phage Display Screening for Anti-A3B scFvs
2.2. Protein Purification
2.3. Protein Binding Studies (SEC, SPR, and BLI)
2.4. Immunofluorescence Microscopy
2.5. X-ray Crystallography
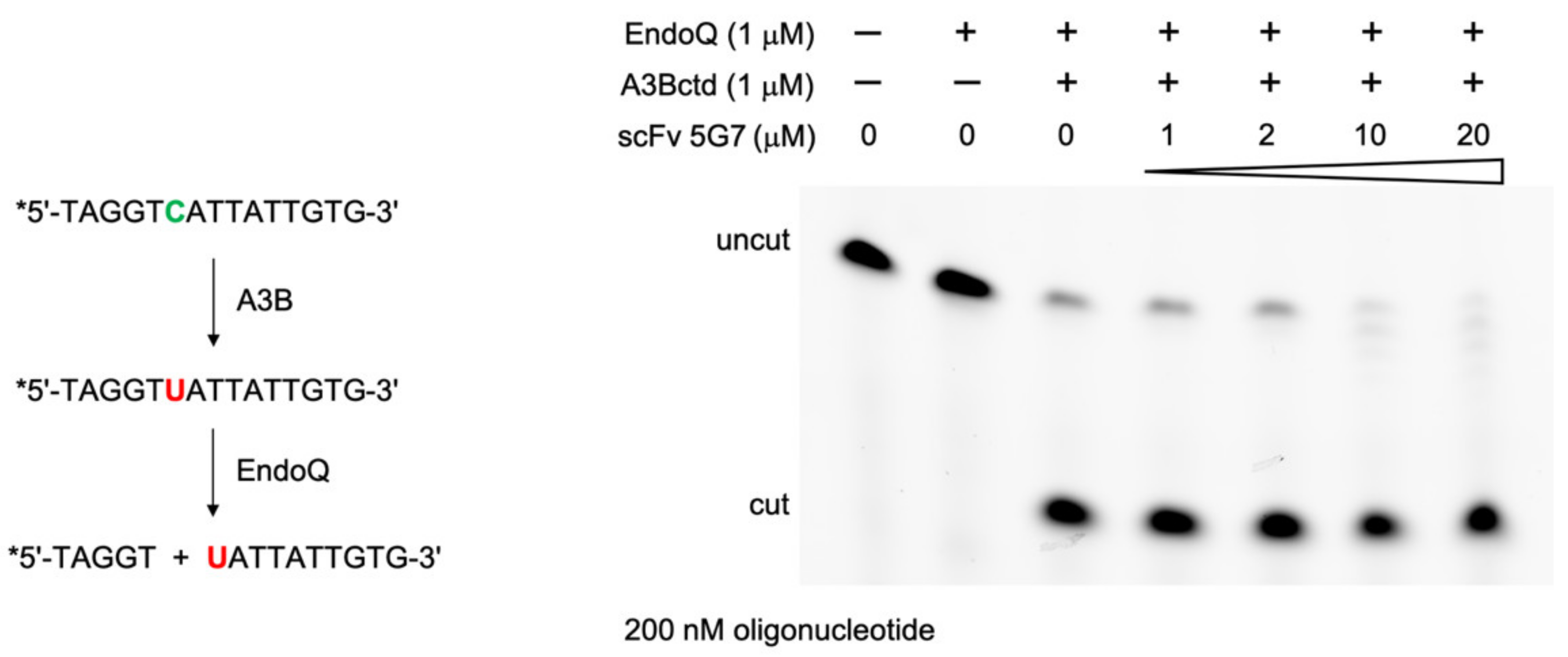
2.6. DNA Deamination Assays
2.7. Computational Docking and MD Simulations
3. Results
3.1. Biopanning Identifies scFV That Binds Human A3B
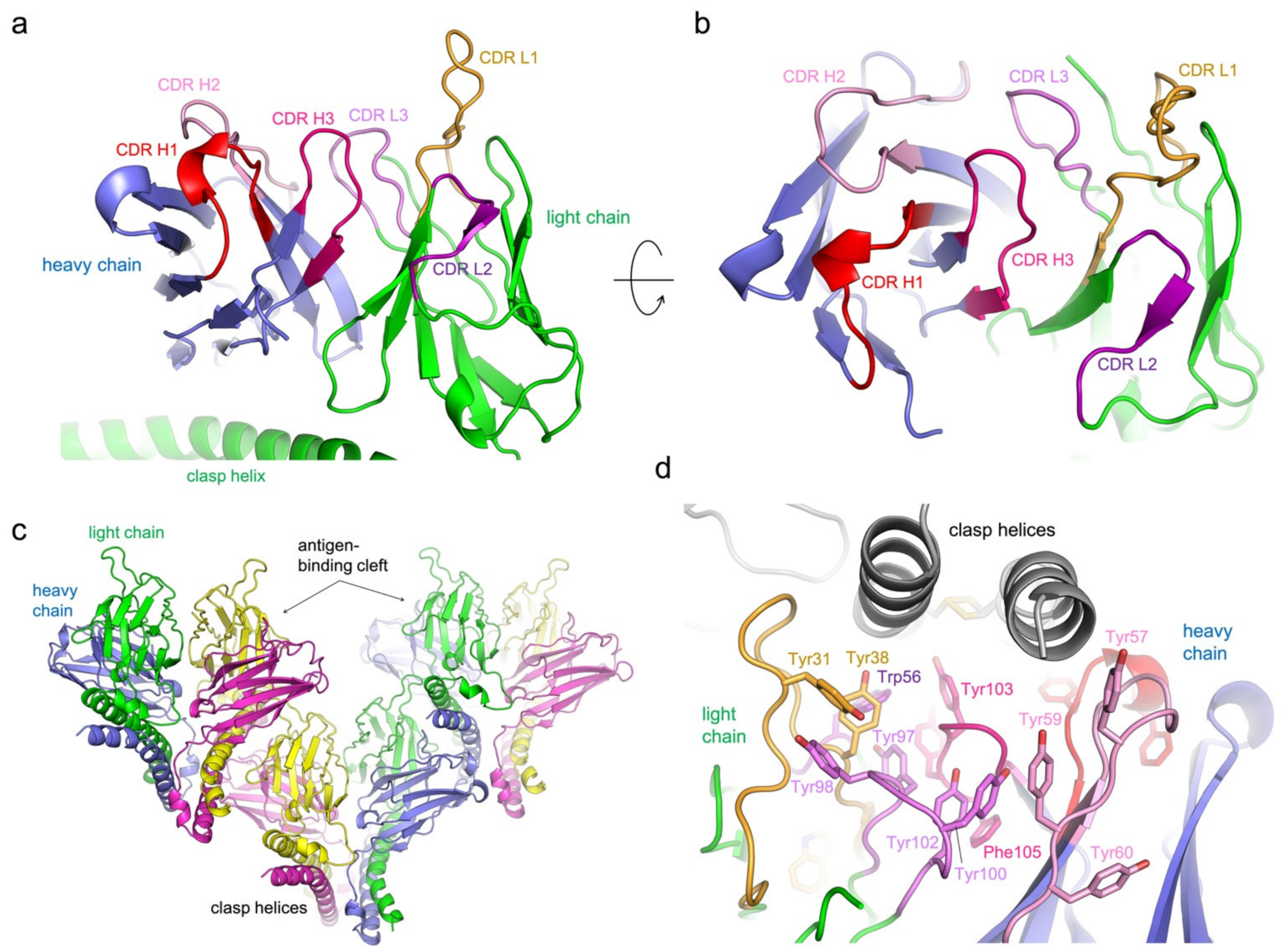
3.2. Structure Determination of 5G7-Clasp
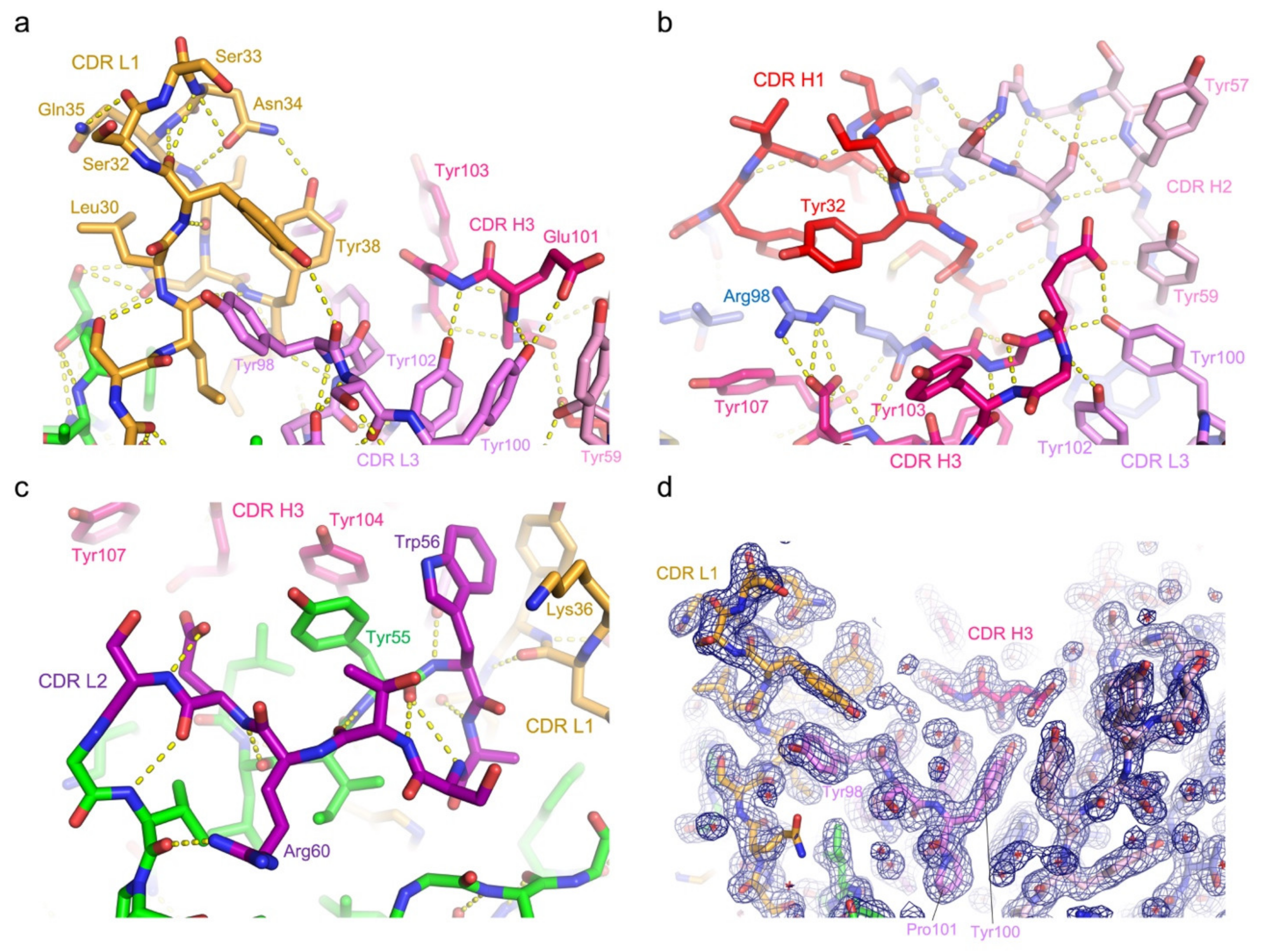
3.3. Protein Interactions in the Antigen-Binding Cleft
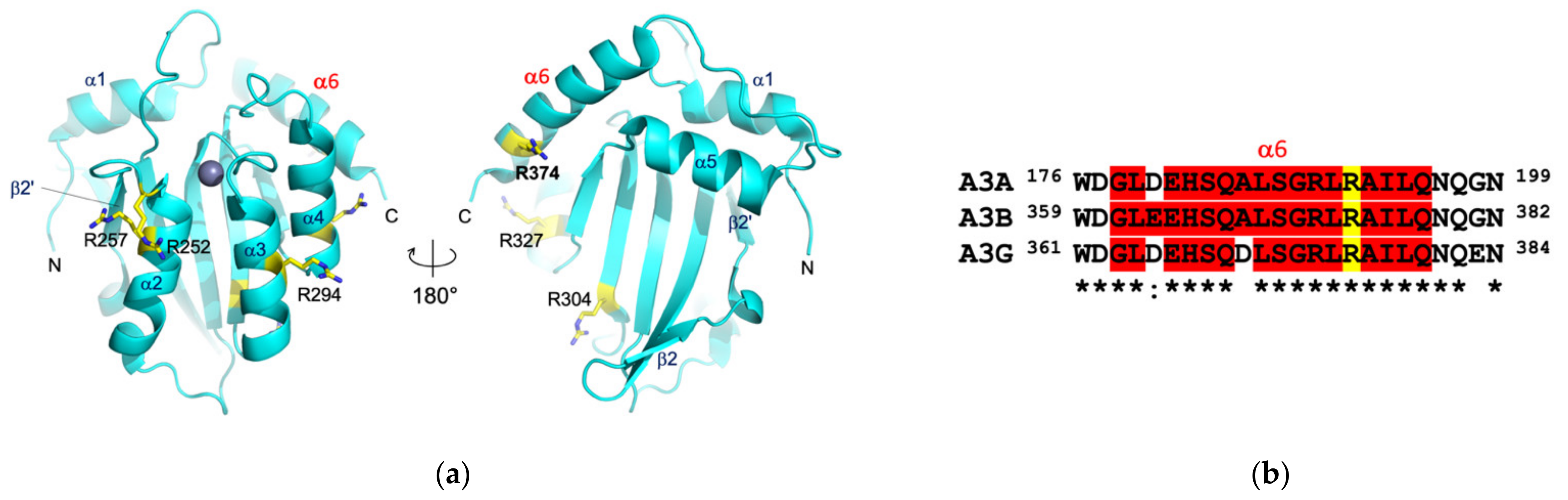
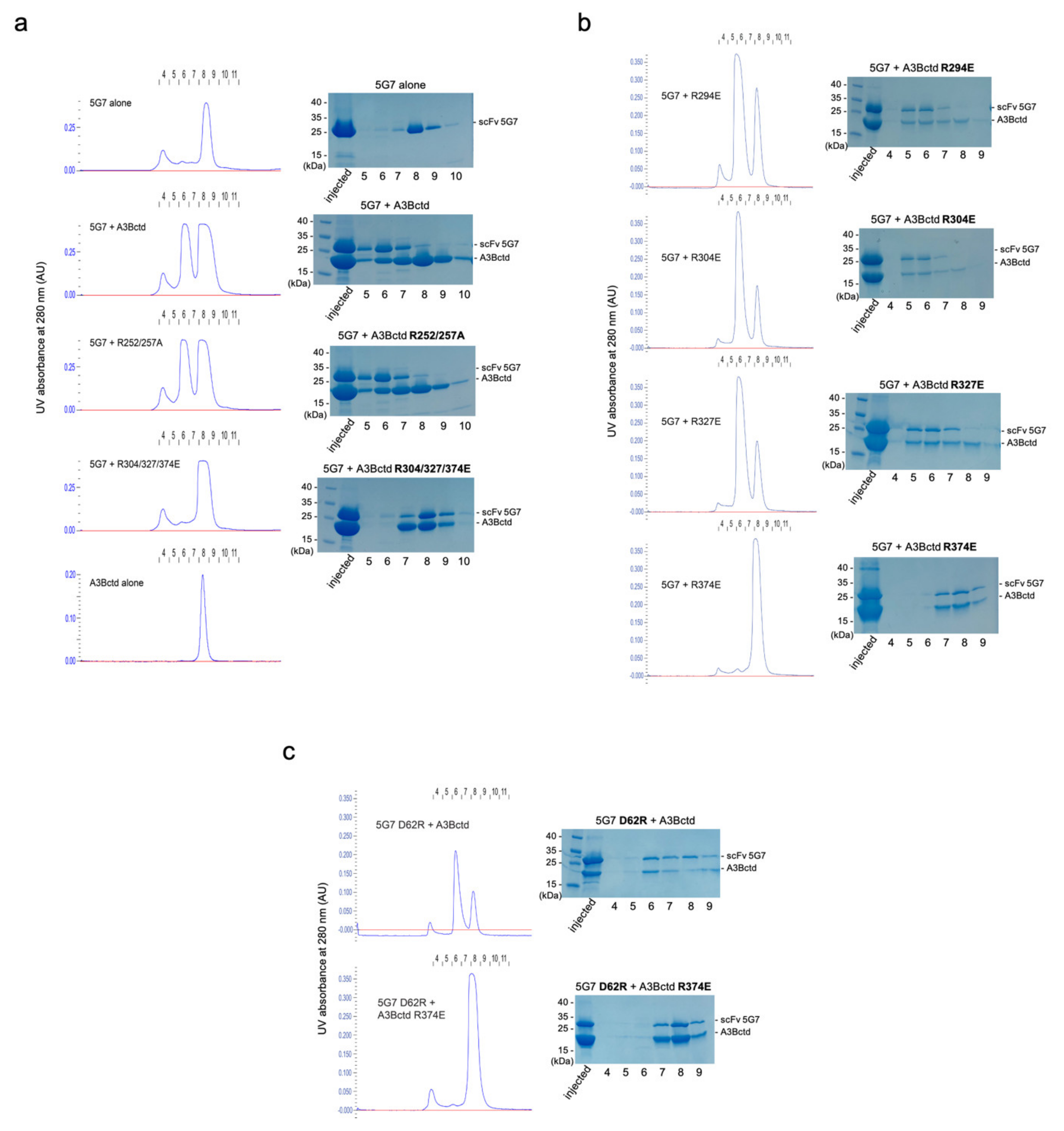
3.4. Epitope Mapping by Site-Directed Mutagenesis
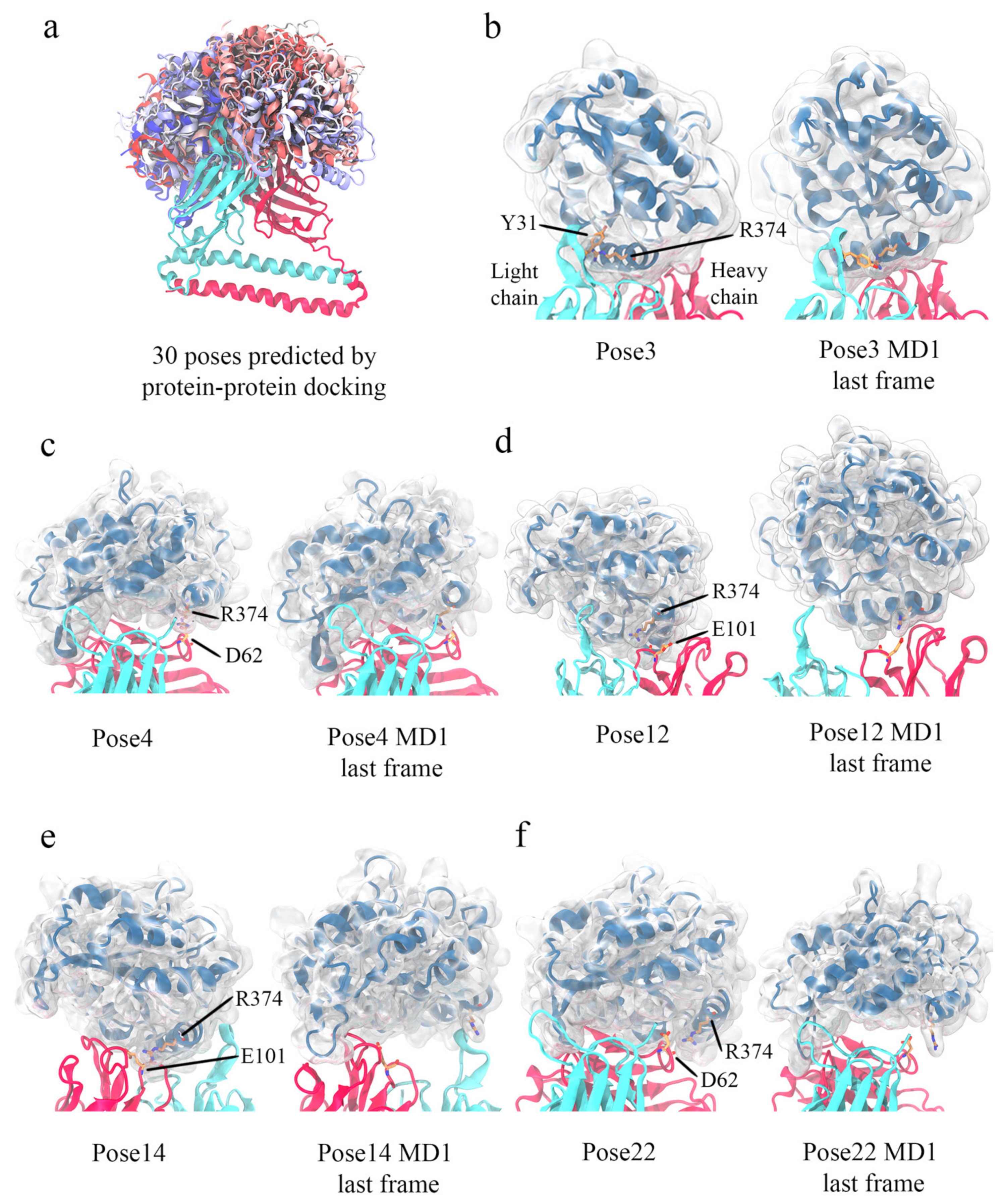
3.5. Computational Simulations of 5G7-A3Bctd Interaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conticello, S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479-480, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siriwardena, S.U.; Chen, K.; Bhagwat, A.S. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 2016, 116, 12688–12710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, A.M.; Weitzman, M.D. The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair 2019, 83, 102700. [Google Scholar] [CrossRef]
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015, 5, 704–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, W.L.; Law, E.K.; Argyris, P.P.; Carpenter, M.A.; Levin-Klein, R.; Ranum, A.N.; Molan, A.M.; Forster, C.L.; Anderson, B.D.; Lackey, L.; et al. A rabbit monoclonal antibody against the antiviral and cancer genomic DNA mutating enzyme APOBEC3B. Antibodies 2019, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Sommavilla, R.; Lovato, V.; Villa, A.; Sgier, D.; Neri, D. Design and construction of a naive mouse antibody phage display library. J. Immunol. Methods 2010, 353, 31–43. [Google Scholar] [CrossRef]
- Shi, K.; Carpenter, M.A.; Kurahashi, K.; Harris, R.S.; Aihara, H. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J. Biol. Chem. 2015, 290, 28120–28130. [Google Scholar] [CrossRef] [Green Version]
- Glumac, P.M.; Forster, C.L.; Zhou, H.; Murugan, P.; Gupta, S.; Lebeau, A.M. The identification of a novel antibody for CD133 using human antibody phage display. Prostate 2018, 78, 981–991. [Google Scholar] [CrossRef]
- Hintz, H.M.; Cowan, A.E.; Shapovalova, M.; Lebeau, A.M. Development of a cross-reactive monoclonal antibody for detecting the tumor stroma. Bioconjug. Chem. 2019, 30, 1466–1476. [Google Scholar] [CrossRef]
- Shi, K.; Demir, O.; Carpenter, M.A.; Banerjee, S.; Harki, D.A.; Amaro, R.E.; Harris, R.S.; Aihara, H. Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B. FASEB BioAdv. 2020, 2, 49–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, K.; Demir, Ö.; Carpenter, M.A.; Wagner, J.; Kurahashi, K.; Harris, R.S.; Amaro, R.E.; Aihara, H. Conformational switch regulates the DNA cytosine deaminase activity of human APOBEC3B. Sci. Rep. 2017, 7, 17415. [Google Scholar] [CrossRef]
- Shi, K.; Carpenter, M.A.; Banerjee, S.; Shaban, N.M.; Kurahashi, K.; Salamango, D.J.; McCann, J.L.; Starrett, G.J.; Duffy, J.V.; Demir, Ö.; et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017, 24, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.-M.; Harjes, E.; Gross, P.J.; Fahmy, A.; Lu, Y.; Shindo, K.; Harris, R.S.; Matsuo, H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 2008, 452, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40 Pt 4, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Arimori, T.; Kitago, Y.; Umitsu, M.; Fujii, Y.; Asaki, R.; Tamura-Kawakami, K.; Takagi, J. Fv-clasp: An artificially designed small antibody fragment with improved production compatibility, stability, and crystallizability. Structure 2017, 25, 1611–1622. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75 Pt 10, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Shi, K.; Moeller, N.H.; Banerjee, S.; McCann, J.L.; Carpenter, M.A.; Yin, L.; Moorthy, R.; Orellana, K.; Harki, D.A.; Harris, R.S.; et al. Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Kozakov, D.; Grove, L.E.; Hall, D.R.; Bohnuud, T.; Mottarella, S.; Luo, L.; Xia, B.; Beglov, D.; Vajda, S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015, 10, 733–755. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, I.; Darden, T.; Duke, R.E.; Giese, T.J.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Schrödinger Release 2019-1: Prime, Schrödinger; LLC: New York, NY, USA, 2019.
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.-P. Novel zinc protein molecular dynamics simulations: Steps toward antiangiogenesis for cancer treatment. J. Mol. Model. 1999, 5, 196–202. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef] [PubMed]
- Beale, R.C.; Petersen-Mahrt, S.K.; Watt, I.N.; Harris, R.S.; Rada, C.; Neuberger, M.S. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: Correlation with mutation spectra in vivo. J. Mol. Biol. 2004, 337, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.; Myint, W.; Kanai, T.; Delviks-Frankenberry, K.; Rodriguez, C.S.; Pathak, V.K.; Schiffer, C.A.; Matsuo, H. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat. Commun. 2018, 9, 2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathore, A.; Carpenter, M.A.; Demir, Ö.; Ikeda, T.; Li, M.; Shaban, N.M.; Law, E.K.; Anokhin, D.; Brown, W.L.; Amaro, R.E.; et al. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 2013, 425, 4442–4454. [Google Scholar] [CrossRef] [Green Version]

| 5G7-Clasp | |
|---|---|
| Data Collection | |
| Wavelength (Å) | 0.979 |
| Resolution range (Å) | 84.9–1.67 (1.73–1.67) |
| Space group | P21 |
| Unit cell | |
| a, b, c (Å) | 54.07 68.82 88.76 |
| α, β, γ (°) | 90 107.05 90 |
| Total reflections | 269,642 (25,086) |
| Unique reflections | 71,734 (7125) |
| Multiplicity | 3.8 (3.5) |
| Completeness (%) | 98.54 (98.81) |
| <I/σ(I)> | 7.88 (1.09) |
| Rmerge | 0.1088 (0.8745) |
| Rmeas | 0.1267 (1.021) |
| Rp.i.m. | 0.06402 (0.518) |
| CC1/2 | 0.997 (0.879) |
| Refinement | |
| Reflections for Rwork | 71,268 (7117) |
| Reflections for Rfree | 3330 (333) |
| Rwork | 0.192 (0.362) |
| Rfree | 0.234 (0.367) |
| No. of non-H atoms | 5935 |
| Macromolecules | 5489 |
| Ligands | 29 |
| Solvent | 417 |
| Protein residues | 672 |
| R.m.s. deviations | |
| Bond length (Å) | 0.006 |
| Bond angles (°) | 0.85 |
| Ramachandran plot | |
| Favored (%) | 98.49 |
| Allowed (%) | 1.51 |
| Outliers (%) | 0.00 |
| Average B-factor | 32.30 |
| Macromolecules | 31.46 |
| Ligands | 52.16 |
| Solvent | 41.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, H.; Demir, Ö.; Kurniawan, F.; Brown, W.L.; Shi, K.; Moeller, N.H.; Carpenter, M.A.; Belica, C.; Orellana, K.; Du, G.; et al. Structural Characterization of a Minimal Antibody against Human APOBEC3B. Viruses 2021, 13, 663. https://doi.org/10.3390/v13040663
Tang H, Demir Ö, Kurniawan F, Brown WL, Shi K, Moeller NH, Carpenter MA, Belica C, Orellana K, Du G, et al. Structural Characterization of a Minimal Antibody against Human APOBEC3B. Viruses. 2021; 13(4):663. https://doi.org/10.3390/v13040663
Chicago/Turabian StyleTang, Heng, Özlem Demir, Fredy Kurniawan, William L. Brown, Ke Shi, Nicholas H. Moeller, Michael A. Carpenter, Christopher Belica, Kayo Orellana, Guocheng Du, and et al. 2021. "Structural Characterization of a Minimal Antibody against Human APOBEC3B" Viruses 13, no. 4: 663. https://doi.org/10.3390/v13040663
APA StyleTang, H., Demir, Ö., Kurniawan, F., Brown, W. L., Shi, K., Moeller, N. H., Carpenter, M. A., Belica, C., Orellana, K., Du, G., LeBeau, A. M., Amaro, R. E., Harris, R. S., & Aihara, H. (2021). Structural Characterization of a Minimal Antibody against Human APOBEC3B. Viruses, 13(4), 663. https://doi.org/10.3390/v13040663






