Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects and Viruses
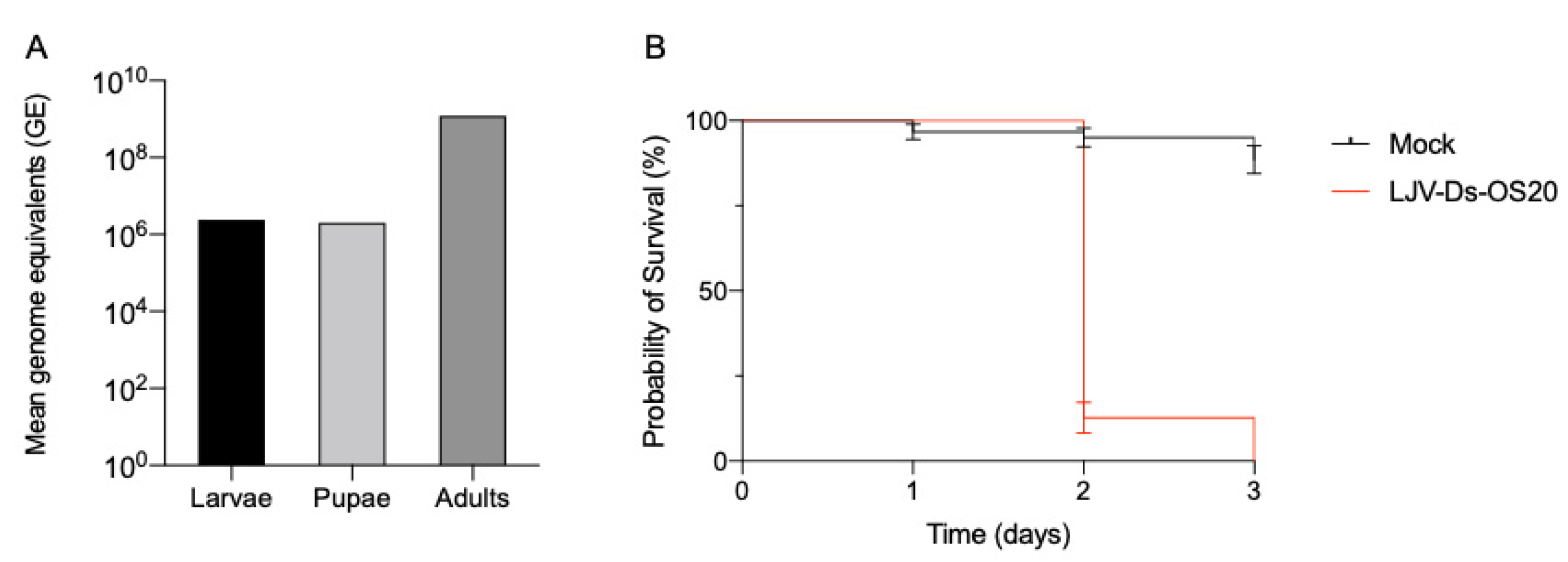
2.2. Virus Infections
2.3. Virus Preparation and Purification
2.4. Transmission Electron Microscopy
2.5. SDS-PAGE, Edman Degradation and Mass Spectrometry
2.6. Quantification of Viral Loads by qRT-PCR
2.7. Sequencing
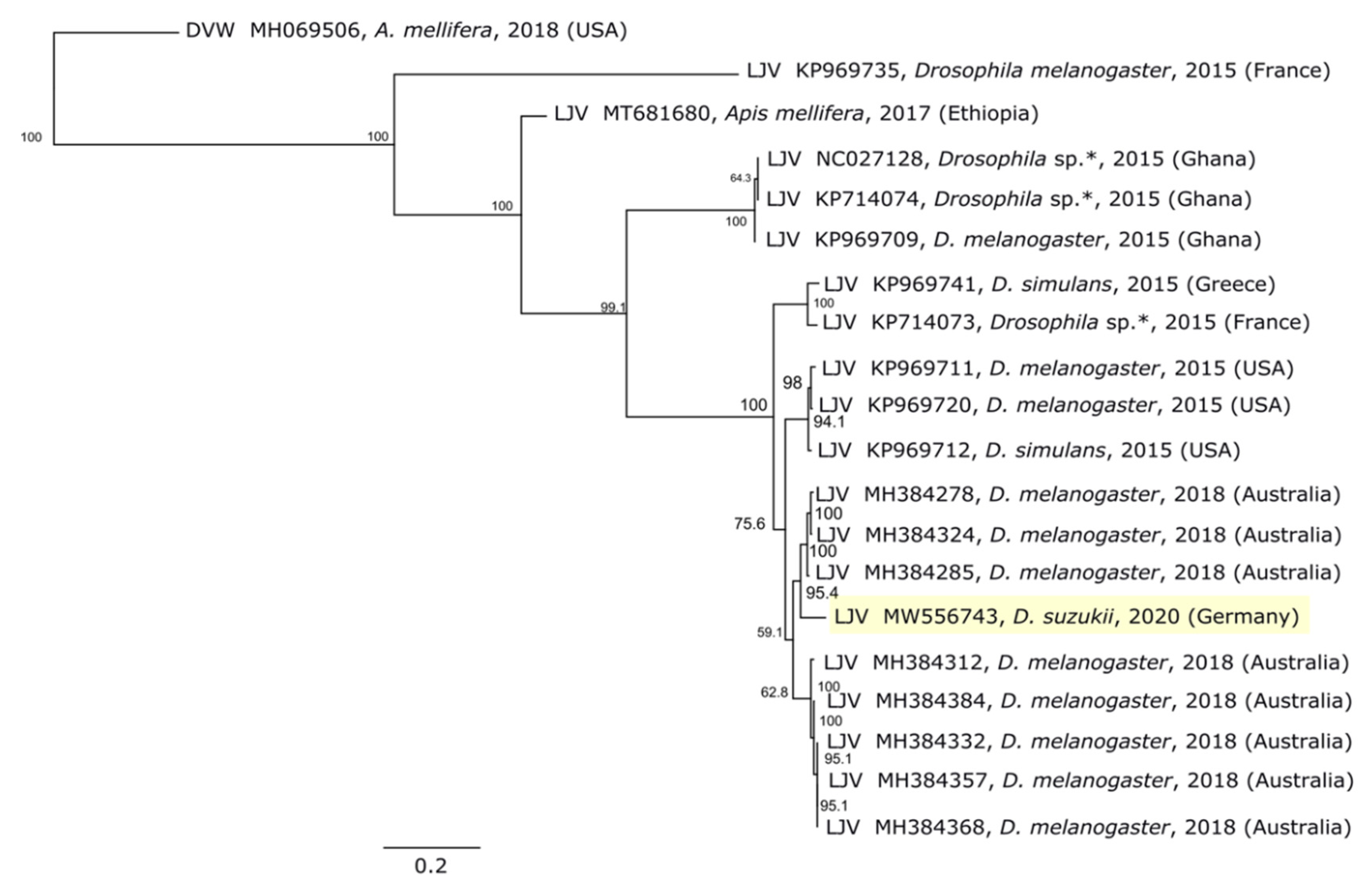
2.8. Phylogenetic Analysis
3. Results
3.1. LJV Production System
3.2. LJV-Ds-OS20 Sequence Analysis
3.3. Purification of LJV-Ds-OS20
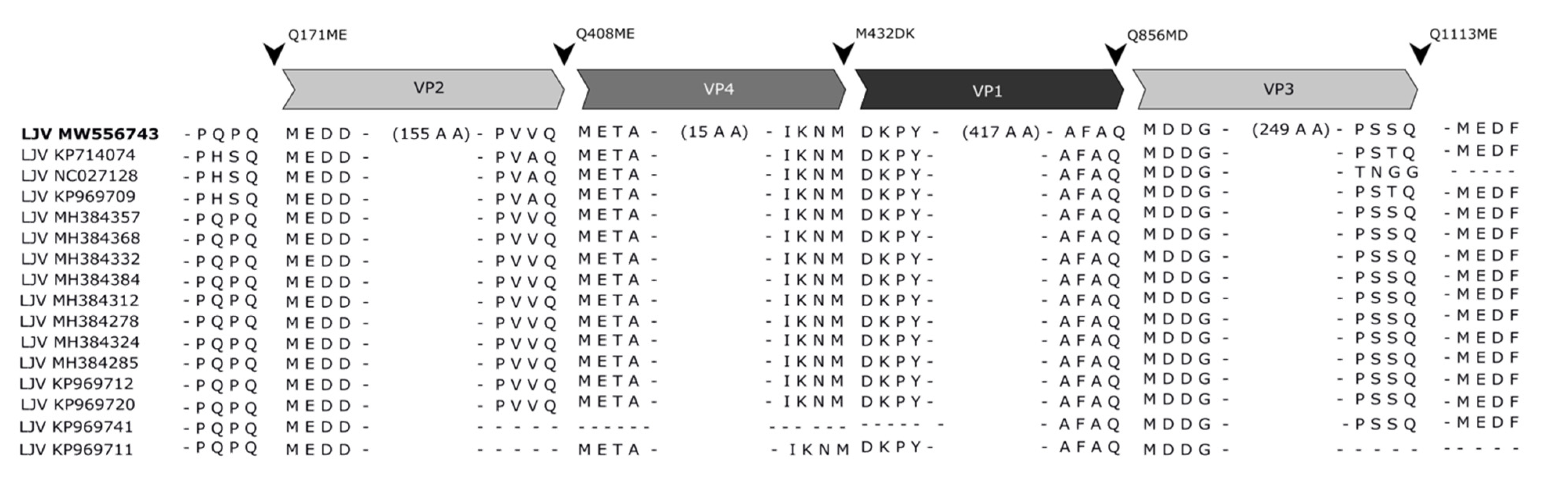
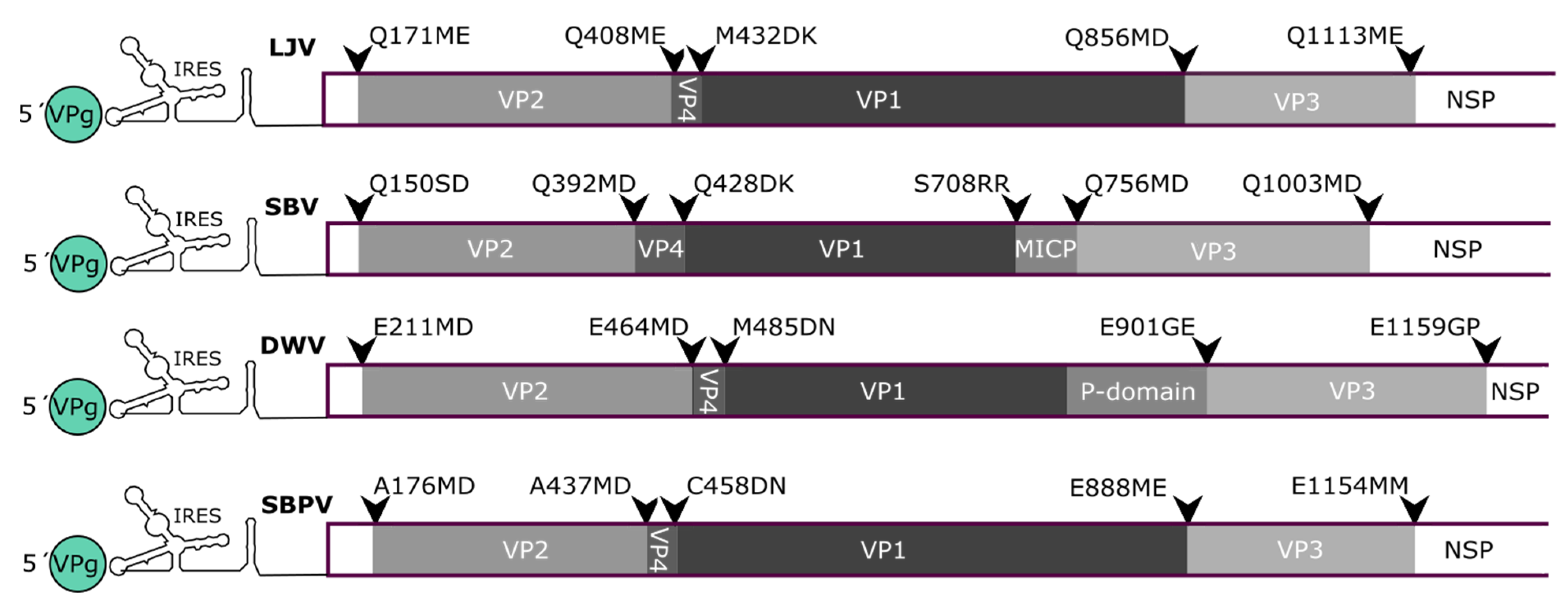
3.4. Mapping of the Structural Protein Region
3.5. Protease Cleavage Sites
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asplen, M.K.; Anfora, G.; Biondi, A.; Choi, D.-S.; Chu, D.; Daane, K.M.; Gibert, P.; Gutierrez, A.P.; Hoelmer, K.A.; Hutchison, W.D.; et al. Invasion biology of Spotted Wing Drosophila (Drosophila suzukii): A Global Perspective and Future Priorities. J. Pest Sci. 2015, 88, 469–494. [Google Scholar] [CrossRef]
- Calabria, G.; Máca, J.; Bächli, G.; Serra, L.; Pascual, M. First records of the potential pest species Drosophila suzukii (Diptera: Drosophilidae) in Europe. J. Appl. Entomol. 2012, 136, 139–147. [Google Scholar] [CrossRef]
- Deprá, M.; Poppe, J.L.; Schmitz, H.J.; Toni, D.C.D.; Valente, V.L.S. The first records of the invasive pest Drosophila suzukii in the South American Continent. J. Pest Sci. 2014, 87, 379–383. [Google Scholar] [CrossRef]
- Walsh, D.B.; Bolda, M.P.; Goodhue, R.E.; Dreves, A.J.; Lee, J.; Bruck, D.J.; Walton, V.M.; O’Neal, S.D.; Zalom, F.G. Drosophila suzukii (Diptera: Drosophilidae): Invasive pest of ripening soft fruit expanding its geographic range and damage potential. J. Integr. Pest Manag. 2011, 2, G1–G7. [Google Scholar] [CrossRef]
- Lee, J.C.; Bruck, D.J.; Curry, H.; Edwards, D.; Haviland, D.R.; Van Steenwyk, R.A.; Yorgey, B.M. The susceptibility of small fruits and cherries to the Spotted-Wing Drosophila, Drosophila suzukii. Pest Manag. Sci. 2011, 67, 1358–1367. [Google Scholar] [CrossRef]
- Atallah, J.; Teixeira, L.; Salazar, R.; Zaragoza, G.; Kopp, A. The Making of a pest: The evolution of a fruit-penetrating ovipositor in Drosophila suzukii and related species. Proc. R. Soc. Lond. B Biol. Sci. 2014, 281, 20132840. [Google Scholar] [CrossRef] [Green Version]
- Tochen, S.; Dalton, D.T.; Wiman, N.; Hamm, C.; Shearer, P.W.; Walton, V.M. Temperature-related development and population parameters for Drosophila suzukii (Diptera: Drosophilidae) on Cherry and Blueberry. Environ. Entomol. 2014, 43, 501–510. [Google Scholar] [CrossRef] [Green Version]
- Wiman, N.G.; Walton, V.M.; Dalton, D.T.; Anfora, G.; Burrack, H.J.; Chiu, J.C.; Daane, K.M.; Grassi, A.; Miller, B.; Tochen, S.; et al. Integrating temperature-dependent life table data into a matrix projection model for Drosophila suzukii population estimation. PLoS ONE 2014, 9, e106909. [Google Scholar] [CrossRef]
- Hunter-Fujita, F.R.; Entwistle, P.F.; Evans, H.F.; Crook, N.E. Insect Viruses and Pest Management; John Wiley & Sons Ltd.: Richmond, Australia, 1998; ISBN 0-471-96878-1. [Google Scholar]
- Lacey, L.A.; Thomson, D.; Vincent, C.; Arthurs, S.P. Codling moth granulovirus: A comprehensive review. Biocontrol Sci. Technol. 2008, 18, 639–663. [Google Scholar] [CrossRef]
- Carrau, T.; Hiebert, N.; Vilcinskas, A.; Lee, K.-Z. Identification and characterization of natural viruses associated with the invasive insect pest Drosophila suzukii. J. Invertebr. Pathol. 2018, 154, 74–78. [Google Scholar] [CrossRef]
- Medd, N.C.; Fellous, S.; Waldron, F.M.; Xuéreb, A.; Nakai, M.; Cross, J.V.; Obbard, D.J. The Virome of Drosophila suzukii, an Invasive Pest of Soft Fruit. Virus Evol. 2018, 4. [Google Scholar] [CrossRef]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E. ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Y.; Sappington, T.W.; Bonning, B.C. Genome sequence of the first coleopteran iflavirus isolated from western corn rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017, 5, e01530-16. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.; Macias-Muñoz, A.; Briscoe, A.D. Genome Sequence of a Novel Iflavirus from MRNA Sequencing of the Butterfly Heliconius erato. Genome Announc. 2014, 2, e00398-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparks, M.E.; Gundersen-Rindal, D.E.; Harrison, R.L. Complete genome sequence of a novel iflavirus from the transcriptome of Halyomorpha halys, the brown marmorated stink bug. Genome Announc. 2013, 1, e00910-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brutscher, L.M.; McMenamin, A.J.; Flenniken, M.L. The buzz about honey bee viruses. PLoS Pathog. 2016, 12, e1005757. [Google Scholar] [CrossRef]
- Aizawa, K.; Kurata, K. Infection under aseptic conditions with virus of infectious flacherie in silkworm Bombyx Mori (Linnaeus). J. Insect Pathol. 1964, 6, 130. [Google Scholar]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and Biological Characterization of Deformed Wing Virus of Honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovac, H.; Crailsheim, K. Lifespan of Apis mellifera Carnica Pollm. Infested by Varroa Jacobsoni Oud. in Relation to Season and Extent of Infestation. J. Apic. Res. 1988, 27, 230–238. [Google Scholar] [CrossRef]
- Škubník, K.; Nováček, J.; Füzik, T.; Přidal, A.; Paxton, R.J.; Plevka, P. Structure of Deformed Wing Virus, a major honey bee pathogen. Proc. Natl. Acad. Sci. USA 2017, 114, 3210–3215. [Google Scholar] [CrossRef] [Green Version]
- Organtini, L.J.; Shingler, K.L.; Ashley, R.E.; Capaldi, E.A.; Durrani, K.; Dryden, K.A.; Makhov, A.M.; Conway, J.F.; Pizzorno, M.C.; Hafenstein, S. Honey Bee Deformed Wing Virus structures reveal that conformational changes accompany genome release. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Procházková, M.; Füzik, T.; Škubník, K.; Moravcová, J.; Ubiparip, Z.; Přidal, A.; Plevka, P. Virion structure and genome delivery mechanism of Sacbrood Honeybee Virus. Proc. Natl. Acad. Sci. USA 2018, 115, 7759–7764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, J.; Saunders, V. Picornaviruses (and other plus-strand RNA viruses). In Virology: Principles and Applications; John Wiley & Sons: Chichester, UK, 2007; pp. 160–165. ISBN 978-0-470-02386-0. [Google Scholar]
- Jakobs, R.; Gariepy, T.D.; Sinclair, B.J. Adult plasticity of cold tolerance in a continental-temperate population of Drosophila suzukii. J. Insect Physiol. 2015, 79, 1–9. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, J.; Liljas, L.; Scotti, P.; Christian, P.; Lin, T.; Johnson, J.E. The Crystal Structure of Cricket Paralysis Virus: The First View of a New Virus Family. Nat. Struct. Biol. 1999, 6, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Reddy, V. Structural studies of nodaviruses and tetraviruses. In The Insect Viruses; Springer Science & Business Media: New York, NY, USA, 1998; pp. 171–223. ISBN 978-1-4615-5341-0. [Google Scholar]
- Schneemann, A.; Reddy, V.; Johnson, J.E. The structure and function of nodavirus particles: A paradigm for understanding chemical biology. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA, 1998; Volume 50, pp. 381–446. [Google Scholar] [CrossRef]
- Hinton, T.M.; Ross-Smith, N.; Warner, S.; Belsham, G.J.; Crabb, B.S. Conservation of L and 3C proteinase activities across distantly related aphthoviruses. J. Gen. Virol. 2002, 83, 3111–3121. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Dainat, B.; Locke, B.; Cordoni, G.; Berthoud, H.; Gauthier, L.; Neumann, P.; Budge, G.E.; Ball, B.V.; Stoltz, D.B. Genetic characterization of Slow Bee Paralysis Virus of the honeybee (Apis mellifera L.). J. Gen. Virol. 2010, 91, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Murakami, R.; Suetsugu, Y.; Nakashima, N. Complete genome sequences of two iflaviruses from the brown planthopper, Nilaparvata lugens. Arch. Virol. 2014, 159, 585–588. [Google Scholar] [CrossRef]
- Grabensteiner, E.; Ritter, W.; Carter, M.J.; Davison, S.; Pechhacker, H.; Kolodziejek, J.; Boecking, O.; Derakhshifar, I.; Moosbeckhofer, R.; Licek, E.; et al. Sacbrood Virus of the Honeybee (Apis mellifera): Rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 2001, 8, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Seitz, K.; Buczolich, K.; Dikunová, A.; Plevka, P.; Power, K.; Rümenapf, T.; Lamp, B. A Molecular clone of Chronic Bee Paralysis Virus (CBPV) causes mortality in honey bee pupae (Apis mellifera). Sci. Rep. 2019, 9, 16274. [Google Scholar] [CrossRef] [Green Version]
- Rueckert, R.R.; Wimmer, E. Systematic nomenclature of picornavirus proteins. J. Virol. 1984, 50, 957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalynych, S.; Füzik, T.; Přidal, A.; de Miranda, J.; Plevka, P. Cryo-EM study of slow bee paralysis virus at low pH reveals Iflavirus genome release mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, 598–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, S.; Xia, H.; Dong, C.; Cheng, Z.; Xia, X.; Zhang, J.; Zhou, X.; Hu, Y. Identification and characterization of Iflavirus 3C-like protease processing activities. Virology 2012, 428, 136–145. [Google Scholar] [CrossRef] [Green Version]
- Kalynych, S.; Přidal, A.; Pálková, L.; Levdansky, Y.; de Miranda, J.R.; Plevka, P. Virion Structure of Iflavirus slow bee paralysis virus at 2.6-Angstrom Resolution. J. Virol. 2016, 90, 7444–7455. [Google Scholar] [CrossRef] [Green Version]
- Farnsworth, D.; Hamby, K.A.; Bolda, M.; Goodhue, R.E.; Williams, J.C.; Zalom, F.G. Economic analysis of revenue losses and control costs associated with the Spotted Wing Drosophila, Drosophila suzukii (Matsumura), in the California Raspberry Industry. Pest Manag. Sci. 2017, 73, 1083–1090. [Google Scholar] [CrossRef] [Green Version]
- Goodhue, R.E.; Bolda, M.; Farnsworth, D.; Williams, J.C.; Zalom, F.G. Spotted Wing Drosophila Infestation of california strawberries and raspberries: Economic analysis of potential revenue losses and control costs. Pest Manag. Sci. 2011, 67, 1396–1402. [Google Scholar] [CrossRef]
- De Ros, G.; Anfora, G.; Grassi, A.; Ioriatti, C. The potential economic impact of Drosophila suzukii on small fruits production in Trentino (Italy). In Proceedings of the IOBC-WPRS Bull, Kusadasi, Turkey, 7 –12 October 2013; Volume 91, pp. 317–321. [Google Scholar]
- Ribière, M.; Olivier, V.; Blanchard, P. Chronic Bee Paralysis: A disease and a virus like no other? J. Invertebr. Pathol. 2010, 103, S120–S131. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Li, P.; Bao, H.; Sun, P.; Bai, X.; Bai, Q.; Li, N.; Ma, X.; Cao, Y.; Fu, Y.; et al. Engineering viable foot-and-mouth disease viruses with increased acid stability facilitate the development of improved vaccines. Appl. Microbiol. Biotechnol. 2020, 104, 1683–1694. [Google Scholar] [CrossRef] [Green Version]



| Oligo Name | FW/RV | Sequence (5′→3′) | Position |
|---|---|---|---|
| LJV_Part_1 | FW | GCTCTGAAGGCCTTGGAAAC | 22–41 |
| LJV_Part_1 | RV | ACGTATCGGGTTCTGTCTCC | 721–702 |
| LJV_Part_2 | FW | GTGTGTGGCGTTACGATTCT | 639–658 |
| LJV_Part_2 | RV | TGCGAGGTCCATCATAACATG | 1334–1314 |
| LJV_Part_3 | FW | TCATACCAAATCGATACACTCCG | 1260–1282 |
| LJV_Part_3 | RV | ATGTACTCAAGAGGCGTCGG | 1960–1941 |
| LJV_Part_4 | FW | GAGGAACCGTGATAGCTGGG | 1913–1932 |
| LJV_Part_4 | RV | ATCTTCGAGTCAGGAGCAGT | 2660–2641 |
| LJV_Part_5 | FW | CCTGGAGCTGTGCAAGTTTC | 2500–2519 |
| LJV_Part_5 | RV | AAAGTAACCGTGCCGCAATT | 3341–3322 |
| LJV_Part_6 | FW | TTCCGATTTTGGCGTGGTTC | 3223–3242 |
| LJV_Part_6 | RV | CGCCCCATGTTTGTAAGCAA | 4019–4000 |
| LJV_Part_7 | FW | TCAACAGGCCTTAACATCACT | 3940–3860 |
| LJV_Part_7 | RV | CTGGCGAACACCTTAAGTCG | 4696–4677 |
| LJV_Part_8 | FW | CGGCGTGTTATTGATCTTCCA | 4585–4560 |
| LJV_Part_8 | RV | TTCGACGTTCTTGGTTGTCA | 5323–5304 |
| LJV_Part_9 | FW | TGAGTGACGAAGAGAGTTTGTC | 5168–5189 |
| LJV_Part_9 | RV | TGGGACATTACAAGGACGGG | 5870–5851 |
| LJV_Part_10 | FW | CCAATCAGTAGGTCCGTGGT | 5802–5821 |
| LJV_Part_10 | RV | TCCGAGTCATTCTGTGCTGT | 6527–6508 |
| LJV_Part_11 | FW | GCACGTATTCCACCTACAGC | 6493–6512 |
| LJV_Part_11 | RV | TCAGGTGACGCTCATTTCCT | 7303–7284 |
| LJV_Part_12 | FW | TCCCGGCATCTCAAAGTGAA | 7199–7218 |
| LJV_Part_12 | RV | AGGTAAGTGCATTTTGGCCG | 7947–7947 |
| LJV_Part_13 | FW | TGAAGTTTCCGGTAAGTGCG | 7869–7888 |
| LJV_Part_13 | RV | AAAGGAGATGCGCAGAACAC | 8576–8557 |
| LJV_Part_14 | FW | TCCTGAAGTTGCGAACCAGA | 8421–8440 |
| LJV_Part_14 | RV | TCGCCGTAAACATACATGCA | 9134–9115 |
| LJV_Part_15 | FW | GGTGAAGGAAAATGGCGTGA | 9079–9098 |
| LJV_Part_15 | RV | TGGATGTGGCACGAAATTACA | 9312–9292 |
| LJV_RACE_1 | RV | GGATTCCAAGAGGTAGTCCCGTGAAC | 241–216 |
| LJV_RACE_2 | RV | CTTTTAGGTGTGGTAGAGTATCATG | 153–129 |
| T1 | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT | Oligo(dT) | |
| T2 | GGCCACGCGTCGACTAGTACGGGGGGGGGGGGGG | Adapter | |
| T22 | GGCCACGCGTCGACTAGTAC | Adapter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrau, T.; Lamp, B.; Reuscher, C.M.; Vilcinskas, A.; Lee, K.-Z. Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii. Viruses 2021, 13, 740. https://doi.org/10.3390/v13050740
Carrau T, Lamp B, Reuscher CM, Vilcinskas A, Lee K-Z. Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii. Viruses. 2021; 13(5):740. https://doi.org/10.3390/v13050740
Chicago/Turabian StyleCarrau, Tessa, Benjamin Lamp, Carina M. Reuscher, Andreas Vilcinskas, and Kwang-Zin Lee. 2021. "Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii" Viruses 13, no. 5: 740. https://doi.org/10.3390/v13050740
APA StyleCarrau, T., Lamp, B., Reuscher, C. M., Vilcinskas, A., & Lee, K.-Z. (2021). Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii. Viruses, 13(5), 740. https://doi.org/10.3390/v13050740







