Utility of Routine Laboratory Biomarkers to Detect COVID-19: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy and Selection Criteria
2.2. Inclusion and Exclusion Criteria
- Published or un-published (pre-print) primary studies or secondary studies where data were used to inform a mathematical model;
- The population included patients suspected of having COVID-19 in hospitals based on their signs and symptoms and where the diagnosis of COVID-19 was confirmed with RT-PCR testing, and radiology;
- Studies reporting the mean (with or without standard deviation) or studies reporting median (with uncertainty measures) of biomarker levels under investigation.
- Analysis of population-based studies with only secondary outcomes such as fatality rate, without discussing the primary outcomes;
- Analysis carried out exclusively on specific sub-populations (elderly, pregnant women, children);
- Case reports and studies with a sample size of less than 10;
- Biomarkers with less than 5 studies were excluded from the meta-analysis;
- Studies reporting biomarker levels, but exclusively for prognostic research or in correlation with severity of disease
- Studies that compare COVID-19-positive patients with controls and patients that have not been tested using RT-PCR (e.g., comparison with other pandemic or with previous flu seasons);
- Studies reporting only the number of patients outside the normal ranges.
2.3. Patient Population
2.4. Study Selection
2.5. Data Extraction and Quality Assessment
2.6. Data Synthesis and Statistical Analysis
3. Results
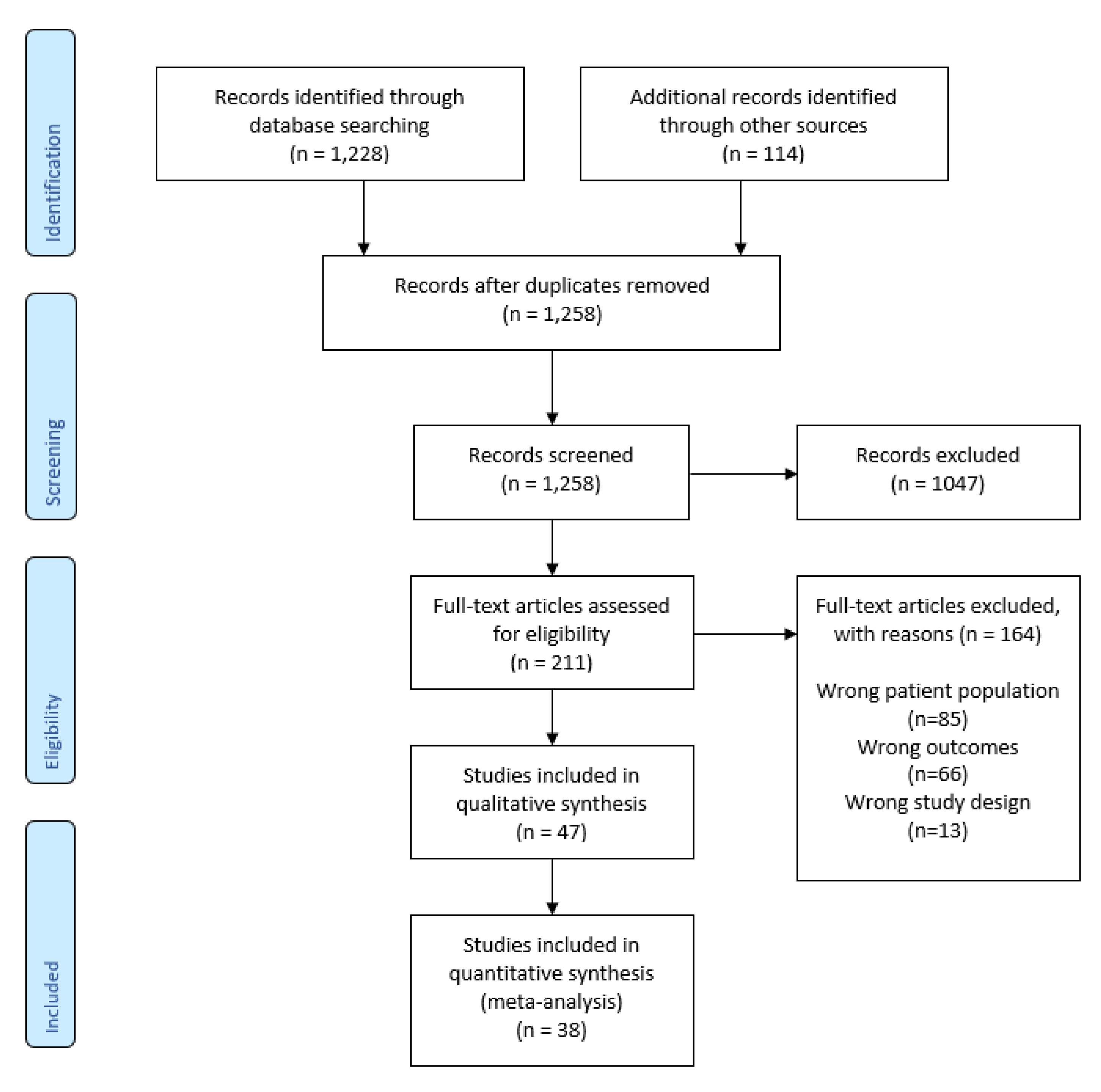
3.1. Literature Retrieval
3.2. Characteristics of Included Studies
3.3. Quality Assessment
3.4. Meta-Analysis
3.5. Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- Population or Problem: Highly suspected and/or confirmed RT-PCR COVID-19-positive patients compared with controls—patients that were sick in the same period of time, but were not diagnosed with COVID-19;
- Intervention or Exposure: diagnosis;
- Context: We are including all studies that report laboratory findings (biomarkers) of hospitalised patients with COVID-19 compared to controls (healthy participants, non-infectious conditions such as bacterial infection) from the same study period;
- Study design: Retrospective studies, primary observational studies, studies where data were used to inform a mathematical model.
- exp Biomarkers/
- (C-reactive protein or CRP) ti.ab.kw.exp C-Reactive Protein/
- Leukocyte ti.ab.kw.exp Leukocytes/
- Lymphocyte.ti,ab,kw.exp Lymphocytes/
- (Lactate Dehydrogenase or LDH).ti,ab,kw.exp L-Lactate Dehydrogenase/
- D-dimer.ti,ab,kw.
- (ferritin or serum ferritin or FRTN).ti,ab,kw.
- (Procalcitonin or PCT).ti,ab,kw.exp Procalcitonin/
- (N-terminal proBNP or NT-proBNP).ti,ab,kw.
- fibrin/ or fibrin fibrinogen degradation products/
- (high-sensitivity cardiac troponin or hs-cTn).ti,ab,kw.Troponin T/Troponin I/
- (brain natriuretic peptide or BNP).ti,ab,kw.Peptide Fragments/ or Natriuretic Peptide, Brain/N-terminal pro-B-type natriuretic peptide.ti,ab,kw.
- exp Eosinophils/Eosinophil count.ti,ab,kw.Eosinophil.ti,ab,kw.
- (Alanine Transaminase or ALT).ti,ab,kw.exp Alanine Transaminases/
- (Aspartate Aminotransferase or AST).ti,ab,kw.exp Aspartate Aminotransferases/
- exp Bilirubin/Bilirubin.ti,ab,kw.
- Laboratory Parameters.ti,ab,kw.Laboratory Findings.ti,ab,kw.Laboratory Results.ti,ab,kw.
- exp coronavirus/
- ((corona* or corono*) adj1 (virus* or viral* or virinae*)).ti,ab,kw.
- (coronavirus* or coronovirus* or coronavirinae* or Coronavirus* or Coronovirus* or Wuhan* or Hubei* or Huanan or “2019-nCoV” or 2019nCoV or nCoV2019 or “nCoV-2019” or “COVID-19” or COVID19 or “CORVID-19” or CORVID19 or “WN-CoV” or WNCoV or “HCoV-19” or HCoV19 or CoV or “2019 novel*” or Ncov or “n-cov” or “SARS-CoV-2” or “SARSCoV-2” or “SARSCoV2” or “SARS-CoV2” or SARSCov19 or “SARS-Cov19” or “SARSCov-19” or “SARS-Cov-19” or Ncovor or Ncorona* or Ncorono* or NcovWuhan* or NcovHubei* or NcovChina* or NcovChinese*).ti,ab,kw.
- (((respiratory* adj2 (symptom* or disease* or illness* or condition*)) or “seafood market*” or “food market*”) adj10 (Wuhan* or Hubei* or China* or Chinese* or Huanan*)).ti,ab,kw.
- ((outbreak* or wildlife* or pandemic* or epidemic*) adj1 (China* or Chinese* or Huanan*)).ti,ab,kw.
- “severe acute respiratory syndrome*”.ti,ab,kw.
- or/1–6
- limit 7 to yr = “2019-Current”
Appendix B
References
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020. [Google Scholar] [CrossRef]
- Singhal, T. A review of coronavirus disease-2019 (COVID-19). Indian J. Pediatr. 2020. [Google Scholar] [CrossRef] [Green Version]
- WHO. Timeline: WHO’s COVID-19 Response. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline#event-14 (accessed on 23 September 2020).
- PHE, COVID-19: Guidance for Sampling and for Diagnostic Laboratories. Available online: https://www.gov.uk/government/publications/wuhan-novel-coronavirus-guidance-for-clinical-diagnostic-laboratories (accessed on 29 July 2020).
- Axell-House, D.B.; Lavingia, R.; Rafferty, M.; Clark, E.; Amirian, E.S.; Chiao, E.Y. The estimation of diagnostic accuracy of tests for COVID-19: A scoping review. J. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Stegeman, I.; Ochodo, E.A.; Guleid, F.; Holtman, G.A.; Yang, B.; Davenport, C.; Deeks, J.J.; Dinnes, J.; Dittrich, S.; Emperador, D. Routine laboratory testing to determine if a patient has COVID-19. Cochrane Database Syst. Rev. 2020. [Google Scholar] [CrossRef]
- Assandri, R.; Montanelli, A. Modified Corona Score can easily identify Covid-19 patients with gastrointestinal symptoms: An Italian proposal. Gastroenterol. Hepatol. Bed Bench 2020, 13, 393–395. [Google Scholar] [PubMed]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.; Debray, T.P.A.; et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- COAP. Living Evidence on COVID-19. Available online: https://zika.ispm.unibe.ch/assets/data/pub/ncov/ (accessed on 23 September 2020).
- Living Overview of the Evidence (L OVE). Available online: https://app.iloveevidence.com/loves/5e6fdb9669c00e4ac072701d (accessed on 23 September 2020).
- QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [CrossRef] [PubMed]
- Hozo, S.P.; Djulbegovic, B.; Hozo, I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res. Methodol. 2005, 5, 13. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.; Welch, V.E. Cochrane Handbook for Systematic Reviews of Interventions Version 6.1; Wiley: Hoboken, NJ, USA, 2019. [Google Scholar]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Review Manager Web (RevMan Web) [Computer Program]. Version 1.22.0. The Cochrane Collaboration. 2020. Available online: revman.cochrane.org (accessed on 3 March 2021).
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- Mardani, R.; Ahmadi Vasmehjani, A.; Zali, F.; Gholami, A.; Mousavi Nasab, S.D.; Kaghazian, H.; Kaviani, M.; Ahmadi, N. Laboratory parameters in detection of COVID-19 patients with positive RT-PCR; a diagnostic accuracy study. Arch. Acad. Emerg. Med. 2020, 8, e43. [Google Scholar] [PubMed]
- Li, Q.; Ding, X.; Xia, G.; Chen, H.-G.; Chen, F.; Geng, Z.; Xu, L.; Lei, S.; Pan, A.; Wang, L.; et al. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: A retrospective case-control study. EClinicalMedicine. 2020, 23. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Ding, F.; Han, L.; Yin, D.; Lu, H.; Zhang, M. The role of peripheral blood eosinophil counts in COVID-19 patients. Allergy 2020, 76, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Kovalic, A.J.; Graber, C.J. Prognostic value of leukocytosis and lymphopenia for coronavirus disease severity. Emerg. Infect. Dis. 2020, 26, 1839–1841. [Google Scholar] [CrossRef]
- Kermali, M.; Khalsa, R.K.; Pillai, K.; Ismail, Z.; Harky, A. The role of biomarkers in diagnosis of COVID-19—A systematic review. Life Sci. 2020, 254, 117788. [Google Scholar] [CrossRef] [PubMed]
- Soraya, G.V.; Ulhaq, Z.S. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: An updated meta-analysis. Med. Clin. 2020, 155, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Ali, N. Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.R.; Kampa, R.K.; Padhi, A.; Panda, A.K. C-reactive protein: A promising biomarker for poor prognosis in COVID-19 infection. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 509, 91–94. [Google Scholar] [CrossRef]
- Gendrel, D.; Bohuon, C. Procalcitonin, a marker of bacterial infection. Infection. 1997, 25, 133–134. [Google Scholar] [CrossRef]
- Gilbert, D.N. Procalcitonin as a biomarker in respiratory tract infection. Clin. Infect. Dis. 2011, 52, S346–S350. [Google Scholar] [CrossRef] [Green Version]
- Vazzana, N.; Dipaola, F.; Ognibene, S. Procalcitonin and secondary bacterial infections in COVID-19: Association with disease severity and outcomes. Acta Clin. Belg. 2020, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.T.; Sun, L.C.; Jia, H.B.; Gao, W.; Yang, J.P.; Zhang, G.Q. Procalcitonin levels in bloodstream infections caused by different sources and species of bacteria. Am. J. Emerg. Med. 2017, 35, 579–583. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Han, C.; Pei, S.; Yin, M.; Chen, X. Procalcitonin levels in COVID-19 patients. Int. J. Antimicrob. Agents 2020, 56, 106051. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chim. Acta 2020, 505, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Graziadio, S.; Hicks, T.; Allen, A.J.; Suklan, J.; Urwin, S.G.; Winter, A.; Price, D.A.; Body, R. A Composite Reference Standard for COVID-19 Diagnostic Accuracy Studies: A Roadmap; PAHO: Washington, DC, USA, 2020. [Google Scholar]
- Rodriguez-Morales, A.J.; Cardona-Ospina, J.A.; Gutiérrez-Ocampo, E.; Villamizar-Peña, R.; Holguin-Rivera, Y.; Escalera-Antezana, J.P.; Alvarado-Arnez, L.E.; Bonilla-Aldana, D.K.; Franco-Paredes, C.; Henao-Martinez, A.F.; et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2020, 101623. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; Lippi, G.; Plebani, M. Laboratory abnormalities in children with novel coronavirus disease 2019. Clin. Chem. Lab. Med. 2020, 20200272. [Google Scholar] [CrossRef] [Green Version]
- Lippi, G.; Plebani, M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin. Chem Lab. Med. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ali, N. Relationship between COVID-19 infection and liver injury: A review of recent data. Front. Med. 2020, 7. [Google Scholar] [CrossRef]
- Gavriatopoulou, M.; Korompoki, E.; Fotiou, D.; Ntanasis-Stathopoulos, I.; Psaltopoulou, T.; Kastritis, E.; Terpos, E.; Dimopoulos, M.A. Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med. 2020, 20, 493–506. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Dramé, M.; Teguo, M.T.; Proye, E.; Hequet, F.; Hentzien, M.; Kanagaratnam, L.; Godaert, L. Should RT-PCR be considered a gold standard in the diagnosis of Covid-19? J. Med. Virol. 2020, 92, 2312–2313. [Google Scholar] [CrossRef] [PubMed]
- Umemneku Chikere, C.M.; Wilson, K.; Graziadio, S.; Vale, L.; Allen, A.J. Diagnostic test evaluation methodology: A systematic review of methods employed to evaluate diagnostic tests in the absence of gold standard—An update. PLoS ONE 2019, 14, e0223832. [Google Scholar] [CrossRef] [PubMed]
- Naaktgeboren, C.A.; Bertens, L.C.M.; Smeden, M.v.; Groot, J.A.H.d.; Moons, K.G.M.; Reitsma, J.B. Value of composite reference standards in diagnostic research. BMJ Br. Med. J. 2013, 347, f5605. [Google Scholar] [CrossRef] [Green Version]
- Testing in UK. Coronavirus (COVID-19) in the UK. GOV.UK. Available online: https://coronavirus.data.gov.uk/details/testing (accessed on 20 April 2021).
- Public Health England. COVID-19: Guidance for The Remobilisation of Services within Health and Care Settings Infection Prevention and Control Recommendations; Public Health England: London, UK, 2020.
- Yao, Y.; Cao, J.; Wang, Q.; Shi, Q.; Liu, K.; Luo, Z.; Chen, X.; Chen, S.; Yu, K.; Huang, Z.; et al. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: A case control study. J. Intensive Care 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, C.F.D.; Queiroz, V.G.S.; Moulin, T.C.; Carvalho, C.A.M.; Haas, C.B.; Rayêe, D.; Henshall, D.E.; De-Souza, E.A.; Amorim, F.E.; Boos, F.Z.; et al. Comparing quality of reporting between preprints and peer-reviewed articles in the biomedical literature. bioRxiv 2020, 581892. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suklan, J.; Cheaveau, J.; Hill, S.; Urwin, S.G.; Green, K.; Winter, A.; Hicks, T.; Boath, A.E.; Kernohan, A.; Price, D.A.; et al. Utility of Routine Laboratory Biomarkers to Detect COVID-19: A Systematic Review and Meta-Analysis. Viruses 2021, 13, 803. https://doi.org/10.3390/v13050803
Suklan J, Cheaveau J, Hill S, Urwin SG, Green K, Winter A, Hicks T, Boath AE, Kernohan A, Price DA, et al. Utility of Routine Laboratory Biomarkers to Detect COVID-19: A Systematic Review and Meta-Analysis. Viruses. 2021; 13(5):803. https://doi.org/10.3390/v13050803
Chicago/Turabian StyleSuklan, Jana, James Cheaveau, Sarah Hill, Samuel G. Urwin, Kile Green, Amanda Winter, Timothy Hicks, Anna E. Boath, Ashleigh Kernohan, D. Ashley Price, and et al. 2021. "Utility of Routine Laboratory Biomarkers to Detect COVID-19: A Systematic Review and Meta-Analysis" Viruses 13, no. 5: 803. https://doi.org/10.3390/v13050803
APA StyleSuklan, J., Cheaveau, J., Hill, S., Urwin, S. G., Green, K., Winter, A., Hicks, T., Boath, A. E., Kernohan, A., Price, D. A., Allen, A. J., Moloney, E., & Graziadio, S. (2021). Utility of Routine Laboratory Biomarkers to Detect COVID-19: A Systematic Review and Meta-Analysis. Viruses, 13(5), 803. https://doi.org/10.3390/v13050803







