The Spike of Concern—The Novel Variants of SARS-CoV-2
Abstract
:1. Introduction
2. The Structure and Biology of Spike
3. The D614G Mutation in Spike Facilitates SARS-CoV-2 Evolution
3.1. The UK Variant B.1.1.7 (VOC202012/01, 20I/501Y.V1)
3.2. The Brazilian P.1 (20J/501Y.V3) Variant
3.3. The North American Variants B.1.429 and B.1.526
3.4. The South African B.1.351 Variant
3.5. The Danish B.1.1.298 Variant
3.6. The Indian B.1.617 Variant
3.7. The Evolution of the New Strains
Funding
Conflicts of Interest
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet Lond. Engl. 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Mercatelli, D.; Giorgi, F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the Numbers. eLife 2020, 9. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef]
- Xiong, C.; Jiang, L.; Chen, Y.; Jiang, Q. Evolution and variation of 2019-novel coronavirus. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- McCallum, M.; Marco, A.D.; Lempp, F.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347. [Google Scholar] [CrossRef]
- Cerutti, G.; Guo, Y.; Zhou, T.; Gorman, J.; Lee, M.; Rapp, M.; Reddem, E.R.; Yu, J.; Bahna, F.; Bimela, J.; et al. Potent SARS-CoV-2 neutralizing antibodies directed against Spike N-terminal domain target a single supersite. Cell Host Microbe 2021, 29, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 183, 1735. [Google Scholar] [CrossRef]
- Lau, S.-Y.; Wang, P.; Mok, B.W.-Y.; Zhang, A.J.; Chu, H.; Lee, A.C.-Y.; Deng, S.; Chen, P.; Chan, K.-H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-NCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by Cryo-EM. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Agajanian, S.; Oztas, D.Y.; Gupta, G. Comparative perturbation-based modeling of the SARS-CoV-2 spike protein binding with host receptor and neutralizing antibodies: Structurally adaptable allosteric communication hotspots define spike sites targeted by global circulating mutations. Biochemistry 2021, 1c00139. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019-NCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Porollo, A.; Meller, J. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinform. 2007, 8, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020, 183, 739–751. [Google Scholar] [CrossRef]
- Hamed, S.M.; Elkhatib, W.F.; Khairalla, A.S.; Noreddin, A.M. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci. Rep. 2021, 11, 8435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Omotuyi, I.O.; Nash, O.; Ajiboye, O.B.; Iwegbulam, C.G.; Oyinloye, E.B.; Oyedeji, O.A.; Kashim, Z.A.; Okaiyeto, K. Atomistic simulation reveals structural mechanisms underlying D614G spike glycoprotein-enhanced fitness in SARS-COV-2. J. Comput. Chem. 2020, 41, 2158–2161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372, eabg0821. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.M.; Moreno, G.K.; Halfmann, P.J.; Hodcroft, E.B.; Baker, D.A.; Boehm, E.C.; Weiler, A.M.; Haj, A.K.; Hatta, M.; Chiba, S.; et al. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathog. 2021, 17, e1009373. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Shi, P.-Y.; Plante, J.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.; Lokugamage, K.; Zhang, X.; Muruato, A.; Zou, J.; et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness and Neutralization Susceptibility. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St. Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383. [Google Scholar] [CrossRef]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 2021, 26. [Google Scholar] [CrossRef]
- Xie, X.; Liu, Y.; Zhang, X.; Zou, J.; Fontes-Garfias, C.; Xia, H.; Swanson, K.; Cutler, M.; Cooper, D.; Menachery, V.; et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621. [Google Scholar] [CrossRef]
- Kustin, T.; Harel, N.; Finkel, U.; Perchik, S.; Harari, S.; Tahor, M.; Caspi, I.; Levy, R.; Leschinsky, M.; Dror, S.K.; et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2 MRNA vaccinated individuals. medRxiv 2021. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Q.; Wei, P.; Chen, Z.; Aviszus, K.; Yang, J.; Downing, W.; Peterson, S.; Jiang, C.; Liang, B.; et al. The basis of a more contagious 501Y.V1 variant of SARS-COV-2. Cell Res. 2021, 1–3. [Google Scholar] [CrossRef]
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Saunders, K.; Edwards, R.J.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. BioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. BioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.-Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Rathnasinghe, R.; Jangra, S.; Cupic, A.; Martínez-Romero, C.; Mulder, L.C.F.; Kehrer, T.; Yildiz, S.; Choi, A.; Mena, I.; De Vrieze, J.; et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. MedRxiv Prepr. Serv. Health Sci. 2021. [Google Scholar] [CrossRef]
- Davies, N.G.; Jarvis, C.I.; CMMID COVID-19 Working Group; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to MRNA vaccine-elicited antibodies. Nature 2021, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 Evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Tang, T.; Daniel, S.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: Role of the P681H mutation. BioRxiv Prepr. Serv. Biol. 2021. [Google Scholar] [CrossRef]
- Detection of New SARS-CoV-2 Variants Related to Mink. 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf (accessed on 5 April 2021).
- Fournier, P.-E.; Colson, P.; Levasseur, A.; Devaux, C.A.; Gautret, P.; Bedotto, M.; Delerce, J.; Brechard, L.; Pinault, L.; Lagier, J.-C.; et al. Emergence and outcome of the SARS-CoV-2 “Marseille-4” variant. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 106, 228–236. [Google Scholar] [CrossRef]
- McCallum, M.; Bassi, J.; Marco, A.D.; Chen, A.; Walls, A.C.; Iulio, J.D.; Tortorici, M.A.; Navarro, M.-J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 Immune Evasion By Variant B.1.427/B.1.429. Biorxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/2021.03.31.437925v1 (accessed on 5 April 2021).
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in california carrying a L452R spike protein mutation. Cell 2021. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Mohri, H.; Zucker, J.E.; Sheng, Z.; Wang, P.; Gomez-Simmonds, A.; Ho, D.D.; Uhlemann, A.-C. A Novel SARS-CoV-2 variant of concern, B.1.526, identified in New York. medRxiv 2021. [Google Scholar] [CrossRef]
- Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings—SARS-CoV-2 Coronavirus/NCoV-2019 Genomic Epidemiology. Available online: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (accessed on 5 April 2021).
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in manaus, brazil. Science 2021, eabh2644. [Google Scholar] [CrossRef]
- Phylogenetic Relationship of SARS-CoV-2 Sequences from Amazonas with Emerging Brazilian Variants Harboring Mutations E484K and N501Y in the Spike Protein—SARS-CoV-2 Coronavirus/NCoV-2019 Genomic Epidemiology. Available online: https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585 (accessed on 5 April 2021).
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Das, S.; Agarwal, A.; Singh, S.; Abraham, P.; et al. Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Available online: https://www.biorxiv.org/content/10.1101/2021.04.22.440932v1 (accessed on 25 April 2021).
- Chen, C.; Boorla, V.S.; Banerjee, D.; Chowdhury, R.; Cavener, V.S.; Nissly, R.H.; Gontu, A.; Boyle, N.R.; Vandergrift, K.; Nair, M.S.; et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike protein and the human ACE2 receptor. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gan, H.H.; Twaddle, A.; Marchand, B.; Gunsalus, K.C. Structural modeling of the SARS-CoV-2 spike/human ACE2 complex interface can identify high-affinity variants associated with increased transmissibility. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ku, Z.; Xie, X.; Davidson, E.; Ye, X.; Su, H.; Menachery, V.D.; Li, Y.; Yuan, Z.; Zhang, X.; Muruato, A.E.; et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021, 12, 469. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef]
- Diamond, M.; Chen, R.; Xie, X.; Case, J.; Zhang, X.; VanBlargan, L.; Liu, Y.; Liu, J.; Errico, J.; Winkler, E.; et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq. 2021, 3, 228079. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.R.; Islam, S.A.; Sternberg, M.J.E. EzMol: A web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J. Mol. Biol. 2018, 430, 2244–2248. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Ikeda, T.; Saito, A.; Tan, T.S.; Ngare, I.; Nasser, H.; Kimura, I.; Uriu, K.; et al. An emerging SARS-CoV-2 mutant evading cellular immunity and increasing viral infectivity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Jumping back and forth: Anthropozoonotic and zoonotic transmission of SARS-CoV-2 on mink farms. Genomics 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.09.01.277152v1.full.pdf (accessed on 11 April 2021).
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361. [Google Scholar] [CrossRef]
- Planas, D.; Bruel, T.; Grzelak, L.; Guivel-Benhassine, F.; Staropoli, I.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 1–8. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Addetia, A.; Hannon, W.W.; Choudhary, M.C.; Dingens, A.S.; Li, J.Z.; Bloom, J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021, 371, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Bayarri-Olmos, R.; Rosbjerg, A.; Johnsen, L.B.; Helgstrand, C.; Bak-Thomsen, T.; Garred, P.; Skjoedt, M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021, 296, 100536. [Google Scholar] [CrossRef] [PubMed]
- Elaswad, A.; Fawzy, M.; Basiouni, S.; Shehata, A.A. Mutational spectra of SARS-CoV-2 isolated from animals. PeerJ 2020, 8, e10609. [Google Scholar] [CrossRef] [PubMed]
- Emergence of SARS-CoV-2 B.1.617 Variants in India and Situation in the EU/EEA. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Emergence-of-SARS-CoV-2-B.1.617-variants-in-India-and-situation-in-the-EUEEA.pdf (accessed on 16 May 2021).
- Yadav, P.D.; Mohandas, S.; Shete, A.M.; Nyayanit, D.A.; Gupta, N.; Patil, D.Y.; Sapkal, G.N.; Potdar, V.; Kadam, M.; Kumar, A.; et al. SARS CoV-2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Sidarovich, A.; Moldenhauer, A.-S.; Winkler, M.S.; Schulz, S.; et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv 2021. [Google Scholar] [CrossRef]
- Avanzato, V.A.; Matson, M.J.; Seifert, S.N.; Pryce, R.; Williamson, B.N.; Anzick, S.L.; Barbian, K.; Judson, S.D.; Fischer, E.R.; Martens, C.; et al. Case study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020, 183, 1901–1912. [Google Scholar] [CrossRef]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K.; et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Hu, F.; Chen, F.; Ou, Z.; Fan, Q.; Tan, X.; Wang, Y.; Pan, Y.; Ke, B.; Li, L.; Guan, Y.; et al. A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract. Cell. Mol. Immunol. 2020, 17, 1119–1125. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 Spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187. [Google Scholar] [CrossRef] [PubMed]
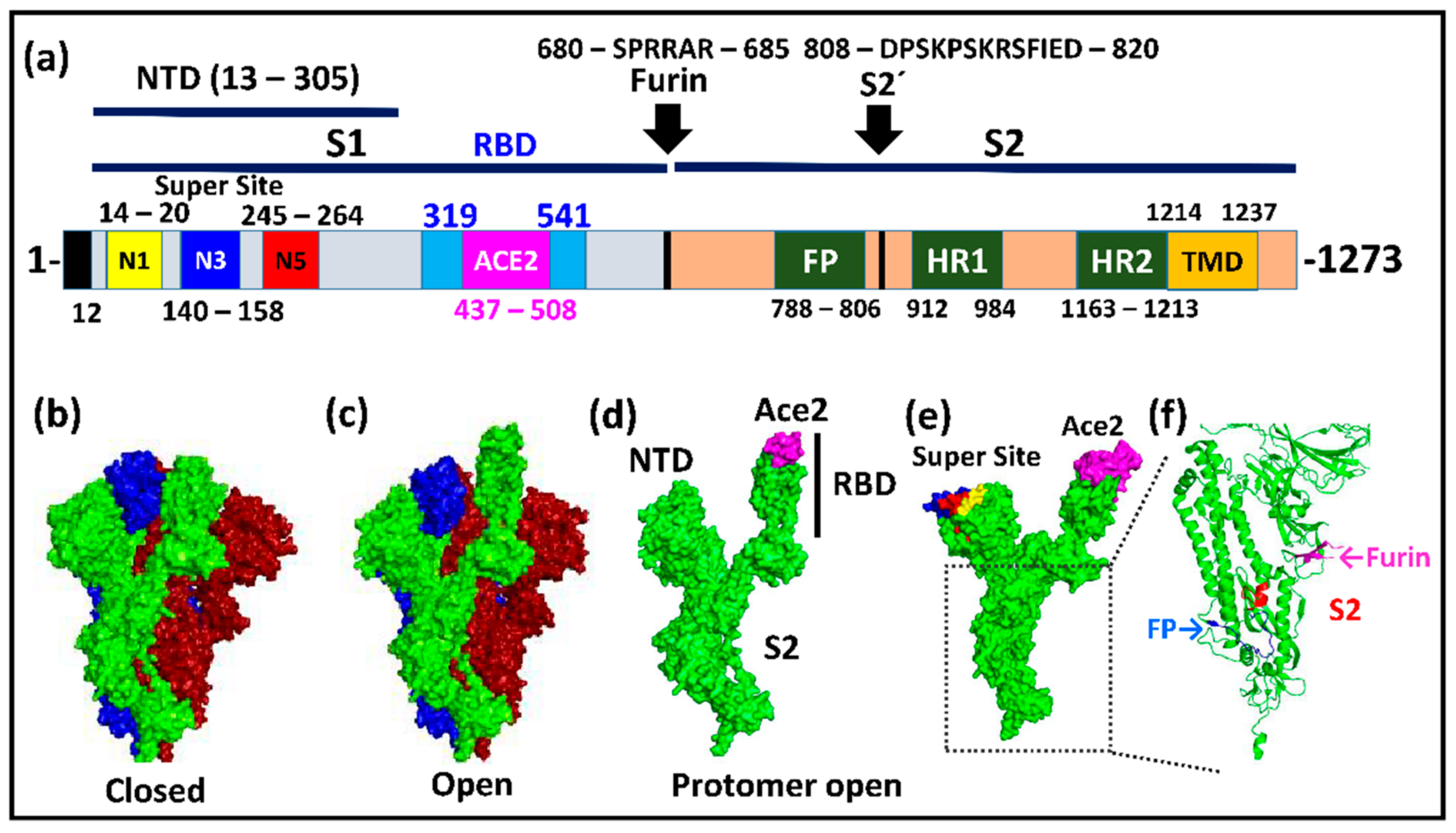
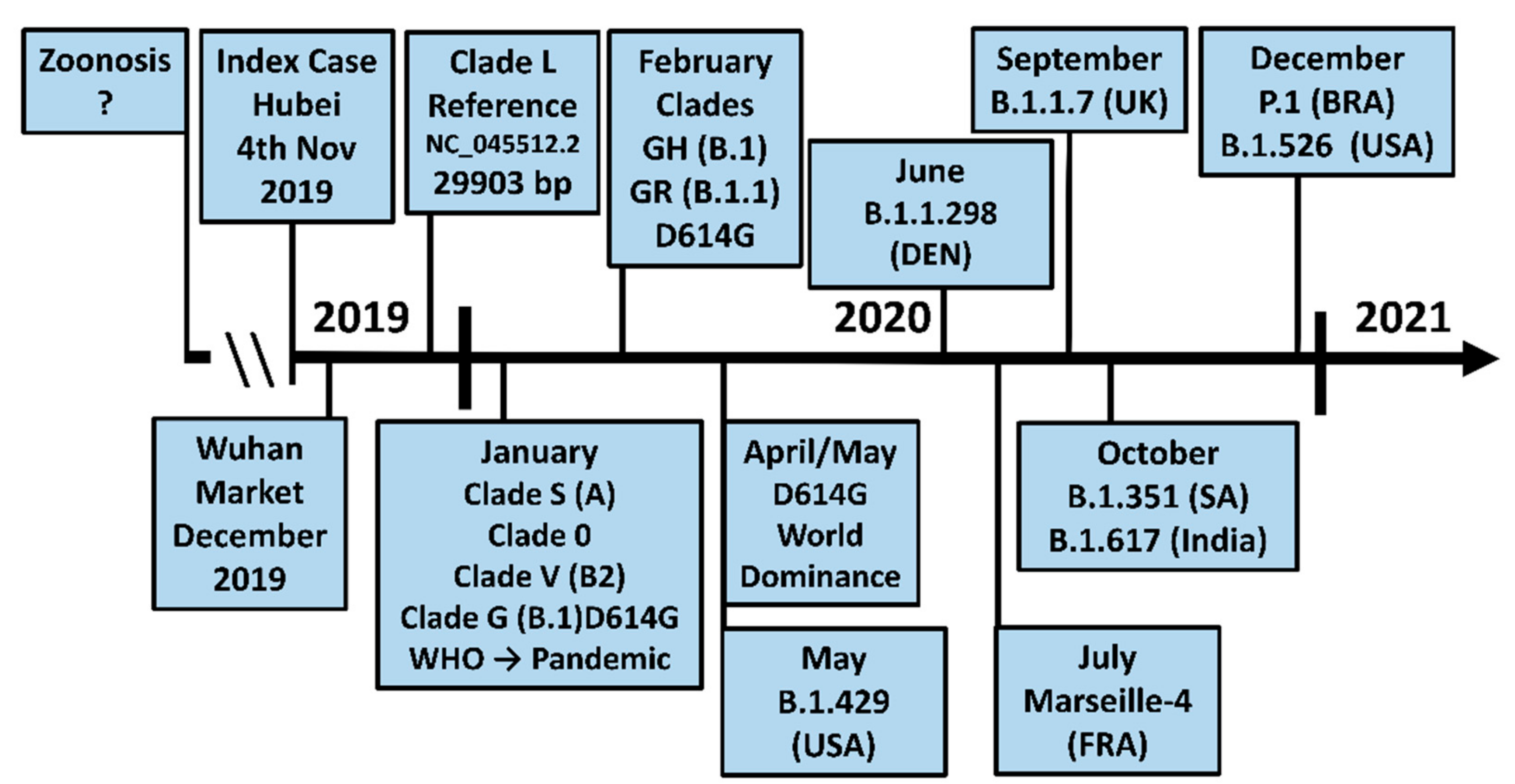


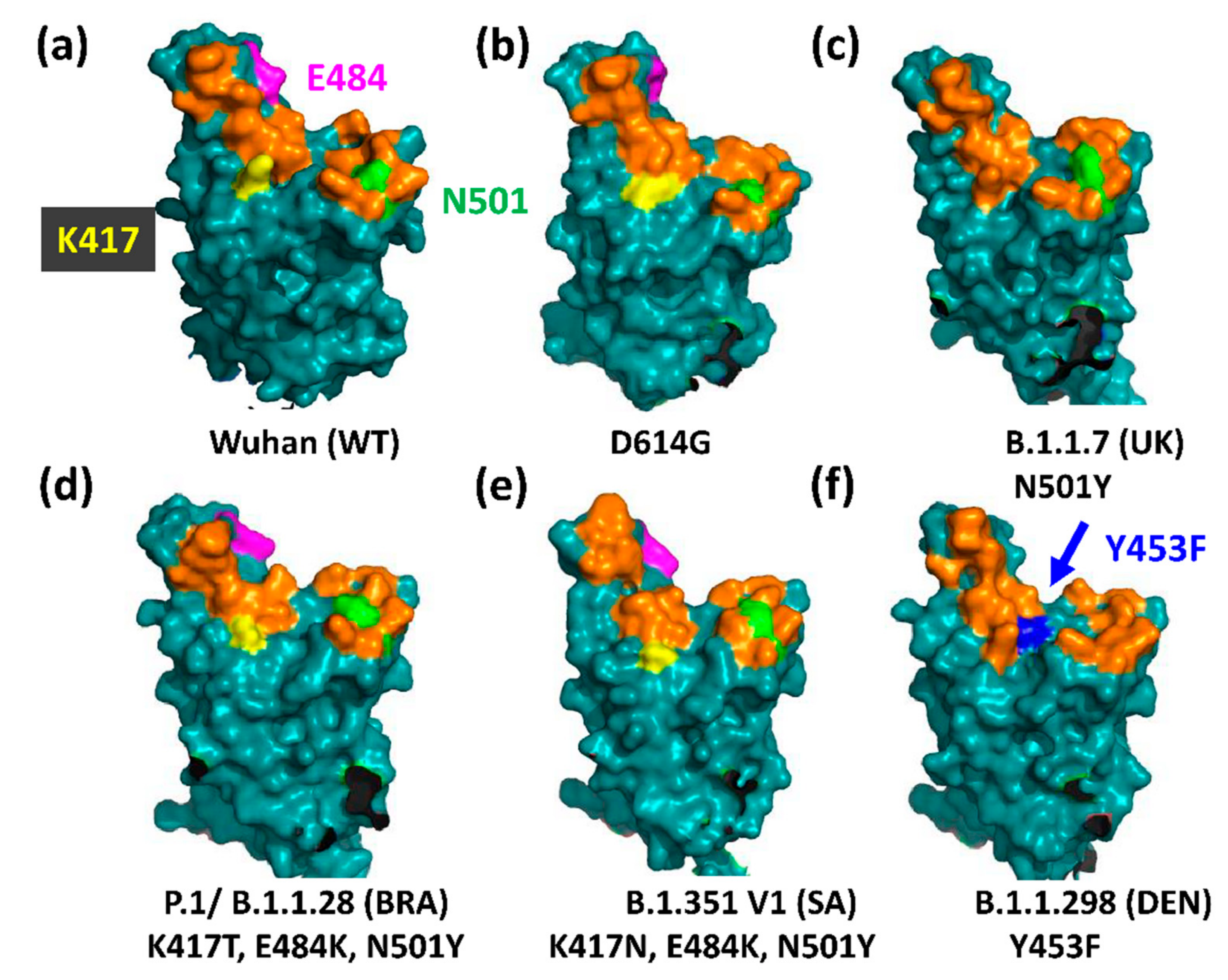


| SARS-CoV-2 Variant | Other S1 Mutations | RBD Mutations | Furin | S2 Mutations |
|---|---|---|---|---|
| B.1.1.7 (UK) [32] | ΔH69, ΔV70, ΔY144, A570D, D614G | N501Y | P681H | T716I, S982A, D1118H |
| B.1.1.298 (DEN) [46] | ΔH69, ΔV70, D614G | Y453F | I692V, M1229I | |
| Marseille-4 (20A.EU2) [47] | S447N | |||
| B.1.429/B.1.427 (USA) [48,49] | S13I, W152C, D614G | L452R | ||
| B.1.526 (USA) [50] | L5F, T95I, D253G, D614G | E484K | A701V | |
| P.1/B.1.1.28 (BRA) [31,51,52] | L18F, T20N, P26S, D138Y, R190S, D614G, H655Y | K417T, E484K, N501Y | T1027I, V1176F * | |
| P.2 (BRA) [53] | D614G | E484K | V1176F | |
| B.1.351 v1 (SA) [54] | D80A, D215G, ΔL242, ΔA243, ΔL244, D614G | K417N, E484K, N501Y | A701V | |
| B.1.351 v2 (SA) [31] | L18F, D80A, D215G, ΔL242, ΔA243, ΔL244, D614G | K417N, E484K, N501Y | A701V | |
| B.1.351 v1 (SA) [31] | D80A, ΔL242, ΔA243, ΔL244, R246I, D614G | K417N, E484K, N501Y | A701V | |
| B.1.617 (India) [55] | G142D, D614G | E484Q, L452R | P681R |
| Replacement | Conformation | hAce2 Affinity | Neutralization | Other |
|---|---|---|---|---|
| S13I | Aberrant cleavage of the signal peptide | |||
| ΔH69, ΔV70 | N2 loop in supersite | Anti-NTD antibodies ↓ | Found in mink | |
| ΔY144 | N3 loop in supersite | Anti-NTD antibodies ↓ | ||
| W152C | Displacement loop N5 | Anti-NTD antibodies ↓ | New disulfide bond C136-C152 | |
| ΔL242, ΔA243, ΔL244 | Displacement loop N5 | Anti-NTD antibodies ↓ | ||
| K417T/N | More RBDs open | Reduced | S1 allosteric centre | |
| L452K | Hydrophobic patchinterrupted | Anti-RBD antibodies ↓ | S1 allosteric centre | |
| Y453F | 4-fold increased | Escape mutation, found in mink | ||
| E484K | More RBDs open | Reduced | Anti-RBD antibodies ↓ | Escape mutation |
| N501Y | More RBDs open | 10-fold increased | Anti-RBD antibodies ↓ | Selected when virus is adapted in mouse, S1 allosteric centre |
| D614G | More RBDs open | Reduced | Not affected | Increased stability |
| P681H/R | Impact on furin cleavage |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winger, A.; Caspari, T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002. https://doi.org/10.3390/v13061002
Winger A, Caspari T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses. 2021; 13(6):1002. https://doi.org/10.3390/v13061002
Chicago/Turabian StyleWinger, Anna, and Thomas Caspari. 2021. "The Spike of Concern—The Novel Variants of SARS-CoV-2" Viruses 13, no. 6: 1002. https://doi.org/10.3390/v13061002
APA StyleWinger, A., & Caspari, T. (2021). The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses, 13(6), 1002. https://doi.org/10.3390/v13061002







