Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions
Abstract
:1. Introduction
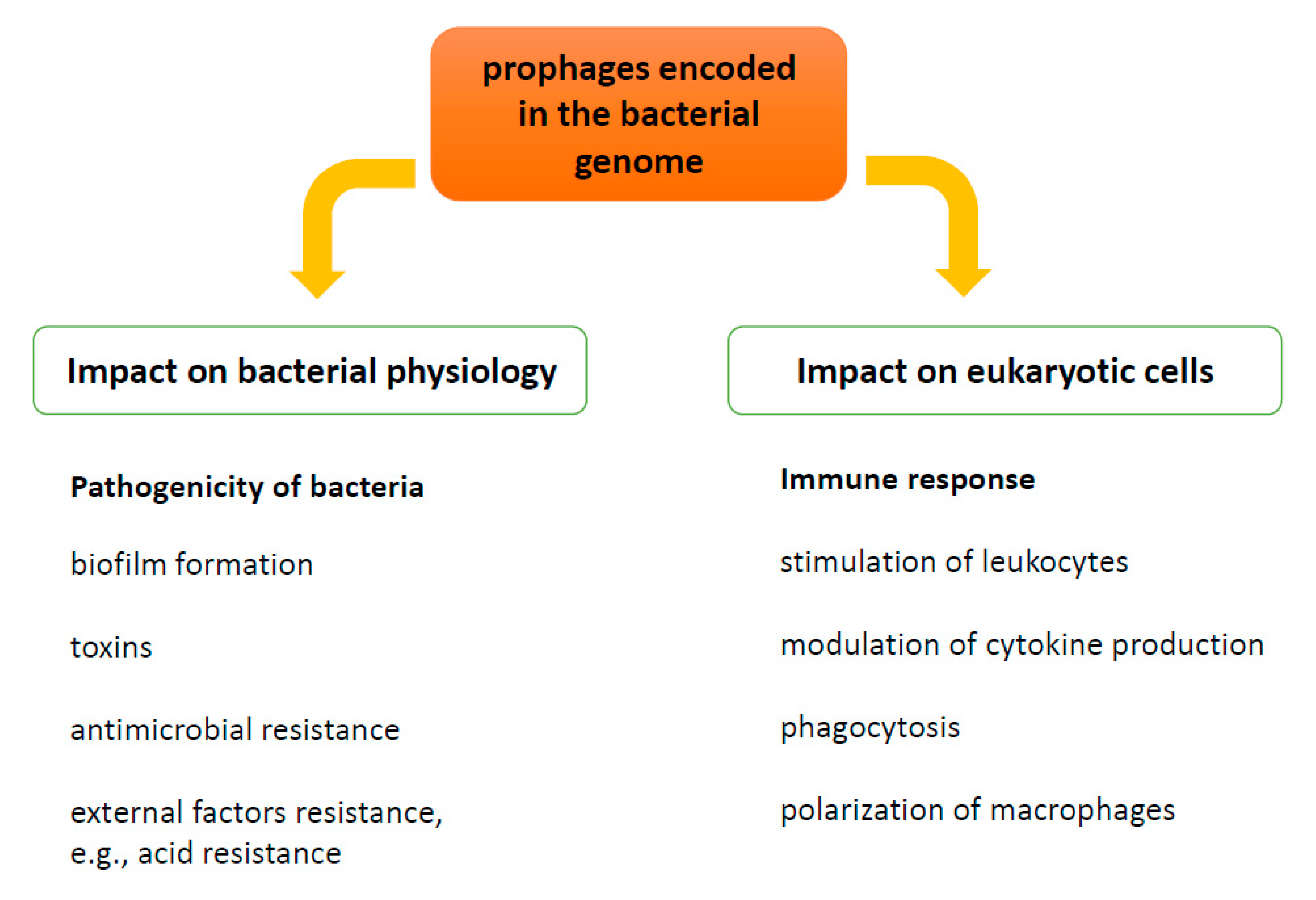
1.1. Prophages as a Modulators of Bacterial Pathogenicity—Benefits for Bacteria
1.1.1. Toxin Production
1.1.2. Biofilm Formation
1.1.3. Antimicrobial Resistance
1.1.4. Bacterial Host Physiology
1.2. Prophages as Modulators of Gut Microbiota
2. Influence of Prophages on Human Immune Response
2.1. Phagocytosis and Intracellular Killing of Bacteria
2.2. Cytokine Production and Function of T-Cells
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lwoff, A. Lysogeny. Bacteriol. Rev. 1953, 17, 269–337. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yang, L.; Yang, D.; Song, J.; Wang, C.; Sun, E.; Gu, C.; Chen, H.; Tong, Y.; Tao, P.; et al. Specific Integration of Temperate Phage Decreases the Pathogenicity of Host Bacteria. Front. Cell Infect. Microbiol. 2020, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Jancheva, M.; Böttcher, T. A Metabolite of Pseudomonas Triggers Prophage-Selective Lysogenic to Lytic Conversion in Staphylococcus aureus. J. Am. Chem. Soc. 2021. [Google Scholar] [CrossRef] [PubMed]
- Harshey, R.M. Transposable Phage Mu. Microbiol. Spectr. 2014, 2, 10. [Google Scholar]
- Casjens, S.R.; Hendrix, R.W. Bacteriophage lambda: Early pioneer and still relevant. Virology 2015, 479-480, 310–330. [Google Scholar] [CrossRef] [Green Version]
- Ravin, N.V. Replication and Maintenance of Linear Phage-Plasmid N15. Microbiol. Spectr. 2015, 3, PLAS-0032-2014. [Google Scholar] [CrossRef] [Green Version]
- Łobocka, M.B.; Rose, D.J.; Plunkett, G., 3rd; Rusin, M.; Samojedny, A.; Lehnherr, H.; Yarmolinsky, M.B.; Blattner, F.R. Genome of bacteriophage P1. J. Bacteriol. 2004, 186, 7032–7068. [Google Scholar] [CrossRef] [Green Version]
- Christie, G.E.; Calendar, R. Bacteriophage P2. Bacteriophage 2016, 6, e1145782. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Renberg, S.K.; Haggård-Ljungquist, E. Derepression of prophage P2 by satellite phage P4: Cloning of the P4 epsilon gene and identification of its product. J. Virol. 1997, 71, 4502–4508. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Renberg, S.K.; Haggård-Ljungquist, E. The E protein of satellite phage P4 acts as an anti-repressor by binding to the C protein of helper phage P2. Mol. Microbiol. 1998, 30, 1041–1050. [Google Scholar] [CrossRef]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and immunomodulation. Future Microbiol. 2017, 12, 905–914. [Google Scholar] [CrossRef] [Green Version]
- Górski, A.; Międzybrodzki, R.; Łobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N.; et al. Phage Therapy: What Have We Learned? Viruses 2018, 10, 288. [Google Scholar] [CrossRef] [Green Version]
- Łobocka, M.; Hejnowicz, M.S.; Gągała, U.; Weber-Dąbrowska, B.; Węgrzyn, G.; Dadlez, M. The first step to bacteriophage therapy—How to choose the correct phage. In Phage Therapy: Current Research and Applications; Borysowski, J., Międzybrodzki, R., Górski, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 23–69. [Google Scholar]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Leitner, L.; Sybesma, W.; Chanishvili, N.; Goderdzishvili, M.; Chkhotua, A.; Ujmajuridze, A.; Schneider, M.P.; Sartori, A.; Mehnert, U.; Bachmann, L.M.; et al. Bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomized, placebo-controlled, double-blind clinical trial. BMC Urol. 2017, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Morozova, V.V.; Vlassov, V.V.; Tikunova, N.V. Applications of Bacteriophages in the Treatment of Localized Infections in Humans. Front. Microbiol. 2018, 9, 1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jończyk-Matysiak, E. The Effect of Bacteriophage Preparations on Intracellular Killing of Bacteria by Phagocytes. Ph.D. Thesis, Ludwik Hirszfeld Institute of Immunology and Experimental Therapy Polish Academy of Sciences, Wrocław, Poland, 2015. [Google Scholar]
- Kurzępa-Skaradzińska, A.; Łusiak-Szelachowska, M.; Skaradziński, G.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Żaczek, M.; Maj, T.; Sławek, A.; Rymowicz, W.; Kłak, M.; et al. Influence of Bacteriophage Preparations on Intracellular Killing of Bacteria by Human Phagocytes in Vitro. Viral. Immunol. 2013, 26, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Kłak, M.; Bubak, B.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Żaczek, M.; Fortuna, W.; Rogóż, P.; Letkiewicz, S.; et al. The Effect of Bacteriophage Preparations on Intracellular Killing of Bacteria by Phagocytes. J. Immunol. Res. 2015, 2015, 482863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Międzybrodzki, R.; Świtała-Jeleń, K.; Fortuna, W.; Weber-Dąbrowska, B.; Przerwa, A.; Łusiak-Szelachowska, M.; Dąbrowska, K.; Kurzępa, A.; Boratyński, J.; Syper, D.; et al. Bacteriophage preparation inhibition of reactive oxygen species generation by endotoxin-stimulated polymorphonuclear leukocytes. Virus Res. 2008, 131, 233–242. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Rouse, M.D.; Stanbro, J.; Roman, J.A.; Lipinski, M.A.; Jacobs, A.; Biswas, B.; Regeimbal, J.; Henry, M.; Stockelman, M.G.; Simons, M.P. Impact of Frequent Administration of Bacteriophage on Therapeutic Efficacy in an A. baumannii Mouse Wound Infection Model. Front. Microbiol. 2020, 11, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miernikiewicz, P.; Dąbrowska, K.; Piotrowicz, A.; Owczarek, B.; Wojas-Turek, J.; Kicielińska, J.; Rossowska, J.; Pajtasz-Piasecka, E.; Hodyra, K.; Macegoniuk, K.; et al. T4 Phage and Its Head Surface Proteins Do Not Stimulate Inflammatory Mediator Production. PLoS ONE 2013, 8, e71036. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef] [Green Version]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral Application of T4 Phage Induces Weak Antibody Production in the Gut and in the Blood. Viruses 2015, 7, 4783–4799. [Google Scholar] [CrossRef] [PubMed]
- Bae, T.; Baba, T.; Hiramatsu, K.; Schneewind, O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 2006, 62, 1035–1047. [Google Scholar] [CrossRef]
- van Wamel, W.J.; Rooijakkers, S.H.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 88, 1310–1315. [Google Scholar] [CrossRef] [Green Version]
- Sela, U.; Euler, C.W.; Correa da Rosa, J.; Fischetti, V.A. Strains of bacterial species induce a greatly varied acute adaptive immune response: The contribution of the accessory genome. PLoS Pathog. 2018, 14, e1006726. [Google Scholar] [CrossRef]
- Fernández, L.; Duarte, A.C.; Rodríguez, A.; García, P. The relationship between the phageome and human health: Are bacteriophages beneficial or harmful microbes? Benef. Microbes 2021, 12, 107–120. [Google Scholar] [CrossRef]
- Bushman, F. Lateral DNA Transfer: Mechanisms and Consequences; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2002. [Google Scholar]
- Bossi, L.; Fuentes, J.A.; Mora, G.; Figueroa-Bossi, N. Prophage contribution to bacterial population dynamics. J. Bacteriol 2003, 185, 6467–6471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlin, G.; Lin, L.; Kudrna, R. Lambda lysogens of E. coli reproduce more rapidly than non-lysogens. Nature 1975, 255, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Edlin, G.; Lin, L.; Bitner, R. Reproductive fitness of P1, P2, and Mu lysogens of Escherichia coli. J. Virol. 1977, 21, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Bitner, R.; Edlin, G. Increased reproductive fitness of Escherichia coli lambda lysogens. J. Virol. 1977, 21, 554–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillol-Salom, A.; Alsaadi, A.; Sousa, J.; Zhong, L.; Foster, K.R.; Rocha, E.; Penadés, J.R.; Ingmer, H.; Haaber, J. Bacteriophages benefit from generalized transduction. PLoS Pathog. 2019, 15, e1007888. [Google Scholar] [CrossRef]
- Burke, J.; Schneider, D.; Westpheling, J. Generalized transduction in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 2001, 98, 6289–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schicklmaier, P.; Schmieger, H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl. Environ. Microbiol. 1995, 61, 1637–1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgson, D.A. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 2000, 35, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F.; Brussow, H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002, 10, 521–529. [Google Scholar] [CrossRef]
- Wagner, P.L.; Waldor, M.K. Bacteriophage control of bacterial virulence. Infect. Immun. 2002, 70, 3985–3993. [Google Scholar] [CrossRef] [Green Version]
- Ptashne, M. A Genetic Switch.: Phage Lambda Revisited, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2004. [Google Scholar]
- Węgrzyn, G.; Węgrzyn, A. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid. Res. Mol. Biol. 2005, 79, 1–48. [Google Scholar]
- Węgrzyn, G.; Licznerska, K.; Węgrzyn, A. Phage lambda--new insights into regulatory circuits. Adv. Virus Res. 2012, 82, 155–178. [Google Scholar] [PubMed]
- Łoś, J.M.; Łoś, M.; Węgrzyn, A.; Węgrzyn, G. Altruism of Shiga toxin-producing Escherichia coli: Recent hypothesis versus experimental results. Front. Cell Infect. Microbiol. 2013, 2, 166. [Google Scholar] [CrossRef] [Green Version]
- Bloch, S.; Nejman-Faleńczyk, B.; Pierzynowska, K.; Piotrowska, E.; Węgrzyn, A.; Marminon, C.; Bouaziz, Z.; Nebois, P.; Jose, J.; Le Borgne, M.; et al. Inhibition of Shiga toxin-converting bacteriophage development by novel antioxidant compounds. J. Enzyme Inhib. Med. Chem. 2018, 33, 639–650. [Google Scholar] [CrossRef] [Green Version]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Davidson, A.R. When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness. J. Microbiol. 2014, 52, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Mai-Prochnow, A.; Hui, J.G.; Kjelleberg, S.; Rakonjac, J.; McDougald, D.; Rice, S.A. Big things in small packages: The genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. 2015, 39, 465–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, L.C. The Contribution of Bacteriophages to the Biology and Virulence of Pathogenic Clostridia. Adv. Appl. Microbiol. 2017, 101, 169–200. [Google Scholar]
- Riedel, T.; Wittmann, J.; Bunk, B.; Schober, I.; Sproer, C.; Gronow, S.; Overmann, J. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J. Biotechnol. 2017, 250, 23–28. [Google Scholar] [CrossRef]
- Jamet, A.; Nassif, X. New players in the toxin field: Polymorphic toxin systems in bacteria. MBio 2015, 6, e00285-15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; de Souza, R.F.; Anantharaman, V.; Iyer, L.M.; Aravind, L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Jamet, A.; Touchon, M.; Ribeiro-Goncalves, B.; Carrico, J.A.; Charbit, A.; Nassif, X.; Ramirez, M.; Rocha, E.P.C. A widespread family of polymorphic toxins encoded by temperate phages. BMC Biol. 2017, 15, 75. [Google Scholar] [CrossRef]
- Kazmi, S.U.; Kansal, R.; Aziz, R.K.; Hooshdaran, M.; Norrby-Teglund, A.; Low, D.E.; Halim, A.-B.; Kotb, M. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group a streptococcal isolates in vivo. Infect. Immun. 2001, 69, 4988–4995. [Google Scholar] [CrossRef] [Green Version]
- Brussow, H.; Hendrix, R.W. Phage genomics: Small is beautiful. Cell 2002, 108, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Benchetrit, L.C.; Gray, E.D.; Wannamaker, L.W. Hyaluronidase activity of bacteriophages of group A streptococci. Infect. Immun. 1977, 15, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Banks, D.J.; Lei, B.; Musser, J.M. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 2003, 71, 7079–7086. [Google Scholar] [CrossRef] [Green Version]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; Jaziri, M.E. The Formation of Biofilms by Pseudomonas aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 759348, 17. [Google Scholar]
- Lasserre, J.F.; Brecx, M.C.; Toma, S. Oral Microbes, Biofilms and Their Role in Periodontal and Peri-Implant Diseases. Materials 2018, 11, 1802. [Google Scholar]
- Colombo, A.P.V.; Tanner, A.C.R. The Role of Bacterial Biofilms in Dental Caries and Periodontal and Peri-implant Diseases: A Historical Perspective. J. Dent. Res. 2019, 98, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, A.A.; Cheng, M.P.; Sheppard, D.C.; Nguyen, D. Microbial Biofilms in Pulmonary and Critical Care Diseases. Ann. Am. Thorac. Soc. 2016, 13, 1615–1623. [Google Scholar] [CrossRef] [Green Version]
- Soto, S.M. Importance of Biofilms in Urinary Tract Infections: New Therapeutic Approaches. Adv. Biol. 2014, 543974, 13. [Google Scholar] [CrossRef]
- Schuch, R.; Fischetti, V.A. The secret life of the anthrax agent Bacillus anthracis: Bacteriophage-mediated ecological adaptations. PLoS ONE 2009, 4, e6532. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001, 413, 860–864. [Google Scholar] [CrossRef]
- Carrolo, M.; Frias, M.J.; Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE 2010, 5, e15678. [Google Scholar] [CrossRef] [Green Version]
- Holt, G.S.; Lodge, J.K.; McCarthy, A.J.; Graham, A.K.; Young, G.; Bridge, S.H.; Brown, A.K.; Veses-Garcia, M.; Lanyon, C.V.; Sails, A.; et al. Shigatoxin encoding Bacteriophage varphi24B modulates bacterial metabolism to raise antimicrobial tolerance. Sci. Rep. 2017, 7, 40424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ubukata, K.; Konno, M.; Fujii, R. Transduction of drug resistance to tetracycline, chloramphenicol, macrolides, lincomycin and clindamycin with phages induced from Streptococcus pyogenes. J. Antibiot. 1975, 28, 681–688. [Google Scholar] [CrossRef] [Green Version]
- Varga, M.; Kuntová, L.; Pantůček, R.; Mašlaňová, I.; Růžičková, V.; Doškař, J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 2012, 332, 146–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, B.; Chen, J.; Manohar, P.; Yu, Y.; Hua, X.; Leptihn, S. A Biological Inventory of Prophages in A. baumannii Genomes Reveal Distinct Distributions in Classes, Length, and Genomic Positions. Front. Microbiol. 2020, 11, 579802. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri Nezhad Fard, R.; Barton, M.D.; Heuzenroeder, M.W. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 2011, 52, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.M.; Veses-Garcia, M.; Hillman, J.D.; Handfield, M.; McCarthy, A.J.; Allison, H.E. Identification of genes expressed in cultures of E. coli lysogens carrying the Shiga toxin-encoding prophage Phi24B. BMC Microbiol. 2012, 12, 42. [Google Scholar] [CrossRef] [Green Version]
- Veses-Garcia, M.; Liu, X.; Rigden, D.J.; Kenny, J.G.; McCarthy, A.J.; Allison, H.E. Transcriptomic analysis of Shiga-toxigenic bacteriophage carriage reveals a profound regulatory effect on acid resistance in Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 8118–8125. [Google Scholar] [CrossRef] [Green Version]
- Flockhart, A.F.; Tree, J.J.; Xu, X.; Karpiyevich, M.; McAteer, S.P.; Rosenblum, R.; Shaw, D.J.; Low, C.J.; Best, A.; Gannon, V.; et al. Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion. Mol. Microbiol. 2012, 83, 208–223. [Google Scholar] [CrossRef] [Green Version]
- Menouni, R.; Hutinet, G.; Petit, M.A.; Ansaldi, M. Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiol. Lett. 2015, 362, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hargreaves, K.R.; Kropinski, A.M.; Clokie, M.R. Bacteriophage behavioral ecology: How phages alter their bacterial host’s habits. Bacteriophage 2014, 4, e29866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duerkop, B.A.; Clements, C.V.; Rollins, D.; Rodrigues, J.L.; Hooper, L.V. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. USA 2012, 109, 17621–17626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanda, A.M.; Thormann, K.; Frunzke, J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 2015, 197, 410–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gama, J.A.; Reis, A.M.; Domingues, I.; Mendes-Soares, H.; Matos, A.M.; Dionisio, F. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS ONE 2013, 8, e59043. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Sausset, R.; Petit, M.A.; Gaboriau-Routhiau, V.; De Paepe, M. New insights into intestinal phages. Mucosal. Immunol. 2020, 13, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Reyes, A.; Haynes, M.; Hanson, N.; Angly, F.E.; Heath, A.C.; Rohwer, F.; Gordon, J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010, 466, 334–338. [Google Scholar] [CrossRef]
- Castro-Mejia, J.L.; Muhammed, M.K.; Kot, W.; Neve, H.; Franz, C.M.; Hansen, L.H.; Vogensen, F.K.; Nielsen, D.S. Optimizing protocols for extraction of bacteriophages prior to metagenomic analyses of phage communities in the human gut. Microbiome 2015, 3, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyles, L.; McCartney, A.L.; Neve, H.; Gibson, G.R.; Sanderson, J.D.; Heller, K.J.; van Sinderen, D. Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 2014, 165, 803–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and their potential to modulate the microbiome and immunity. Cell Mol. Immunol. 2021, 18, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, S.G.; Shamash, M.; Hynes, A.P.; Maurice, C.F. Common Oral Medications Lead to Prophage Induction in Bacterial Isolates from the Human Gut. Viruses 2021, 13, 455. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Turroni, F.; Tremblay, D.; Ferrario, C.; Mancabelli, L.; Duranti, S.; Ward, D.V.; Ossiprandi, M.C.; Moineau, S.; et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ. Microbiol. 2016, 18, 2196–2213. [Google Scholar] [CrossRef] [PubMed]
- Guigas, C.; Faulhaber, K.; Duerbeck, D.; Neve, H.; Heller, K.J. Prophage-mediated modulation of interaction of Streptococcus thermophilus J34 with human intestinal epithelial cells and its competition against human pathogens. Benef. Microbes 2016, 7, 289–297. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Turroni, F.; Duranti, S.; Ferrario, C.; Viappiani, A.; Mancabelli, L.; Mangifesta, M.; Taminiau, B.; Delcenserie, V.; et al. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl. Environ. Microbiol. 2014, 80, 6383–6394. [Google Scholar] [CrossRef] [Green Version]
- Lugli, G.A.; Milani, C.; Turroni, F.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Ferrario, C.; Modesto, M.; Mattarelli, P.; Jiří, K.; et al. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genom. 2017, 18, 568. [Google Scholar] [CrossRef] [Green Version]
- Ventura, M.; Turroni, F.; Lima-Mendez, G.; Foroni, E.; Zomer, A.; Duranti, S.; Giubellini, V.; Bottacini, F.; Horvath, P.; Barrangou, R.; et al. Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl. Environ. Microbiol. 2009, 75, 6929–6936. [Google Scholar] [CrossRef] [Green Version]
- Breitbart, M.; Haynes, M.; Kelley, S.; Angly, F.; Edwards, R.A.; Felts, B.; Mahaffy, J.M.; Mueller, J.; Nulton, J.; Rayhawk, S.; et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008, 159, 367–373. [Google Scholar] [CrossRef]
- Lim, E.S.; Zhou, Y.; Zhao, G.; Bauer, I.K.; Droit, L.; Ndao, I.M.; Warner, B.B.; Tarr, P.I.; Wang, D.; Holtz, L.R. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 2015, 21, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Sharon, I.; Morowitz, M.J.; Thomas, B.C.; Costello, E.K.; Relman, D.A.; Banfield, J.F. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013, 23, 111–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornuault, J.K.; Petit, M.A.; Mariadassou, M.; Benevides, L.; Moncaut, E.; Langella, P.; Sokol, H.; De Paepe, M. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome 2018, 6, 65. [Google Scholar] [CrossRef]
- Sinha, A.; Maurice, C.F. Bacteriophages: Uncharacterized and Dynamic Regulators of the Immune System. Mediat. Inflamm. 2019, 2019, 3730519. [Google Scholar] [CrossRef] [PubMed]
- Metchnikoff, E. Immunity in Infective Diseases; Johnson Reprint Corp: New York, NY, USA, 1968. [Google Scholar]
- Hampton, M.B.; Kettle, A.J.; Winterbourn, C.C. Inside the Neutrophil Phagosome: Oxidants, Myeloperoxidase, and Bacterial Killing. Blood 1998, 92, 3007–3017. [Google Scholar] [CrossRef]
- Kaźmierczak, Z.; Szostak-Paluch, K.; Przybyło, M.; Langner, M.; Witkiewicz, W.; Jędruchniewicz, N.; Dąbrowska, K. Endocytosis in cellular uptake of drug delivery vectors: Molecular aspects in drug development. Bioorg. Med. Chem. 2020, 28, 115556. [Google Scholar] [CrossRef]
- Młynarczyk, G.; Garliński, P.; Młynarczyk, A.; Zabuska, K.; Sawicka-Grzelak, A.; Machowska, G.; Osowiecki, H.; Roszkowski, W. Bacteriophage conversion as a factor modifying the intensity of phagocytosis of Staphylococcus aureus by human leukocytes. Med. Dosw. Mikrobiol. 1989, 41, 86–91. [Google Scholar]
- Młynarczyk, G.; Garliński, P.; Młynarczyk, A.; Zabuska-Jabłońska, K.; Machowska, G.; Sawicka-Grzelak, A.; Roszkowski, W. Increase of pathogenicity Staphylococcus aureus strains caused by lysogenic conversion by phages of serologic group F. Med. Dosw. Mikrobiol. 1993, 45, 19–23. [Google Scholar]
- Folsom, J.P.; Richards, L.; Pitts, B.; Roe, F.; Ehrlich, G.D.; Parker, A.; Mazurie, A.; Stewart, P.S. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 2010, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Secor, P.R.; Michaels, L.A.; Smigiel, K.S.; Rohani, M.G.; Jennings, L.K.; Hisert, K.B.; Arrigoni, A.; Braun, K.R.; Birkland, T.P.; Lai, Y.; et al. Filamentous Bacteriophage Produced by Pseudomonas aeruginosa Alters the Inflammatory Response and Promotes Noninvasive Infection In Vivo. Infect. Immun. 2016, 85, e00648-16. [Google Scholar] [CrossRef] [Green Version]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front. Immunol. 2020, 11, 244. [Google Scholar] [CrossRef] [Green Version]
- Vaca-Pacheco, S.; Paniagua-Contreras, G.L.; García-González, O.; de la Garza, M. The Clinically Isolated FIZ15 Bacteriophage Causes Lysogenic Conversion in Pseudomonas aeruginosa PAO1. Curr. Microbiol. 1999, 38, 239–243. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363, eaat9691. [Google Scholar] [CrossRef]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasechnek, A.; Rabinovich, L.; Stadnyuk, O.; Azulay, G.; Mioduser, J.; Argov, T.; Borovok, I.; Sigal, N.; Herskovits, A.A. Active Lysogeny in Listeria Monocytogenes Is a Bacteria-Phage Adaptive Response in the Mammalian Environment. Cell Rep. 2020, 32, 107956. [Google Scholar] [CrossRef]
- Lackie, J. A Dictionary of Biomedicine, 1st ed.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, D.J.; Beres, S.B.; Musser, J.M. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002, 10, 515–521. [Google Scholar] [CrossRef]
- Proft, T.; Moffatt, S.L.; Berkahn, C.J.; Fraser, J.D. Identification and characterization of novel superantigens from Streptococcus pyogenes. J. Exp. Med. 1999, 189, 89–102. [Google Scholar] [CrossRef] [Green Version]
- Bohach, G.A.; Fast, D.J.; Nelson, R.D.; Schlievert, P.M. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit. Rev. Microbiol. 1990, 17, 251–272. [Google Scholar] [CrossRef]
- Kotzin, B.L.; Leung, D.Y.; Kappler, J.; Marrack, P. Superantigens and their potential role in human disease. Adv. Immunol. 1993, 54, 99–166. [Google Scholar] [PubMed]
- Baba, T.; Bae, T.; Schneewind, O.; Takeuchi, F.; Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008, 190, 300–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa-Bossi, N.; Uzzau, S.; Maloriol, D.; Bossi, L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 2001, 39, 260–271. [Google Scholar] [CrossRef]
- Tarr, P.I.; Schoening, L.M.; Yea, Y.L.; Ward, T.R.; Jelacic, S.; Whittam, T.S. Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J. Bacteriol. 2000, 182, 6183–6191. [Google Scholar] [CrossRef] [Green Version]
- Plunkett, G., 3rd; Rose, D.J.; Durfee, T.J.; Blattner, F.R. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 1999, 181, 1767–1778. [Google Scholar] [CrossRef] [Green Version]
- Makino, K.; Yokoyama, K.; Kubota, Y.; Yutsudo, C.H.; Kimura, S.; Kurokawa, K.; Ishii, K.; Hattori, M.; Tatsuno, I.; Abe, H.; et al. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 1999, 74, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Nietfeldt, J.; Ju, J.; Wise, J.; Fegan, N.; Desmarchelier, P.; Benson, A.K. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J. Bacteriol. 2001, 183, 6885–6897. [Google Scholar] [CrossRef] [Green Version]
- Coleman, D.; Knights, J.; Russell, R.; Shanley, D.; Birkbeck, T.H.; Dougan, G.; Charles, I. Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage phi 13 occurs by site- and orientation-specific integration of the phi 13 genome. Mol. Microbiol. 1991, 5, 933–939. [Google Scholar] [CrossRef]
- de Haas, C.J.; Veldkamp, K.E.; Peschel, A.; Weerkamp, F.; Van Wamel, W.J.; Heezius, E.C.; Poppelier, M.J.; Van Kessel, K.P.; van Strijp, J.A. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004, 199, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Dohlsten, M.; Bjorklund, M.; Sundstedt, A.; Hedlund, G.; Samson, D.; Kalland, T. Immunopharmacology of the superantigen staphylococcal enterotoxin A in T-cell receptor V beta 3 transgenic mice. Immunology 1993, 79, 520–527. [Google Scholar]
- Rooijakkers, S.H.; Ruyken, M.; Roos, A.; Daha, M.R.; Presanis, J.S.; Sim, R.B.; van Wamel, W.J.; van Kessel, K.P.; van Strijp, J.A. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005, 6, 920–927. [Google Scholar] [CrossRef]
- Haas, P.J.; de Haas, C.J.; Kleibeuker, W.; Poppelier, M.J.; van Kessel, K.P.; Kruijtzer, J.A.; Liskamp, R.M.; van Strijp, J.A. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. J. Immunol. 2004, 173, 5704–5711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postma, B.; Kleibeuker, W.; Poppelier, M.J.; Boonstra, M.; Van Kessel, K.P.; Van Strijp, J.A.; de Haas, C.J. Residues 10-18 within the C5a receptor N terminus compose a binding domain for chemotaxis inhibitory protein of Staphylococcus aureus. J. Biol. Chem. 2005, 280, 2020–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postma, B.; Poppelier, M.J.; van Galen, J.C.; Prossnitz, E.R.; van Strijp, J.A.; de Haas, C.J.; van Kessel, K.P. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol. 2004, 172, 6994–7001. [Google Scholar] [CrossRef] [Green Version]
- Jin, T.; Bokarewa, M.; Foster, T.; Mitchell, J.; Higgins, J.; Tarkowski, A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 2004, 172, 1169–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooijakkers, S.H.; van Wamel, W.J.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005, 7, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Rahimpour, R.; Mitchell, G.; Khandaker, M.H.; Kong, C.; Singh, B.; Xu, L.; Ochi, A.; Feldman, R.D.; Pickering, J.G.; Gill, B.M.; et al. Bacterial superantigens induce down-modulation of CC chemokine responsiveness in human monocytes via an alternative chemokine ligand-independent mechanism. J. Immunol. 1999, 162, 2299–2307. [Google Scholar] [PubMed]
- Genestier, A.L.; Michallet, M.C.; Prévost, G.; Bellot, G.; Chalabreysse, L.; Peyrol, S.; Thivolet, F.; Etienne, J.; Lina, G.; Vallette, F.M.; et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J. Clin. Investig. 2005, 115, 3117–3127. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Cheng, H.; Yuan, W.; Zeng, F.; Shang, W.; Tang, D.; Xue, W.; Fu, J.; Zhou, R.; Zhu, J.; et al. Panton-Valentine leukocidin (PVL)-positive health care-associated methicillin-resistant Staphylococcus aureus isolates are associated with skin and soft tissue infections and colonized mainly by infective PVL-encoding bacteriophages. J. Clin. Microbiol. 2015, 53, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, D.; Gieldon, L.; Mysore, V.; Nippe, N.; Taxman, D.J.; Duncan, J.A.; Broglie, P.M.; Marketon, K.; Austermann, J.; Vogl, T.; et al. Staphylococcus aureus Panton-Valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J. Leukoc. Biol. 2012, 92, 1069–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoong, P.; Pier, G.B. Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infect. Immun. 2012, 80, 2894–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popescu, M.; Van Belleghem, J.D.; Khosravi, A.; Bollyky, P.L. Bacteriophages and the Immune System. Annu. Rev. Virol. 2021, 8, 67–87. [Google Scholar] [CrossRef] [PubMed]
| Modulation of | Direction of Change |
|---|---|
| T cell proliferation | ↑/↓ [30] |
| B cell proliferation | ↑ [30] |
| IgG synthesis | ↑ [30] |
| IFN-γ or IFN type 1 synthesis | ↑ [30,109] |
| ROS production by leukocytes | ↓ [103,104] |
| levels of leukocytes in the infected tissue (based on the presence of CD45 molecule) | ↓ [106] |
| intracellular killing of bacteria (phagocytosis) | ↓ [103,104,109] |
| macrophage polarization | towards M2 type [106] |
| pro-inflammatory cytokines (e.g., TNF-α, IL-1β, IL-2) in vivo in the infected tissue and in vitro | ↓ [106,109] /↑ [117,118] |
| anti-inflammatory cytokines (e.g., IL-10) in vivo in the infected tissue | ↑ [106] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cieślik, M.; Bagińska, N.; Jończyk-Matysiak, E.; Węgrzyn, A.; Węgrzyn, G.; Górski, A. Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses 2021, 13, 1013. https://doi.org/10.3390/v13061013
Cieślik M, Bagińska N, Jończyk-Matysiak E, Węgrzyn A, Węgrzyn G, Górski A. Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses. 2021; 13(6):1013. https://doi.org/10.3390/v13061013
Chicago/Turabian StyleCieślik, Martyna, Natalia Bagińska, Ewa Jończyk-Matysiak, Alicja Węgrzyn, Grzegorz Węgrzyn, and Andrzej Górski. 2021. "Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions" Viruses 13, no. 6: 1013. https://doi.org/10.3390/v13061013
APA StyleCieślik, M., Bagińska, N., Jończyk-Matysiak, E., Węgrzyn, A., Węgrzyn, G., & Górski, A. (2021). Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses, 13(6), 1013. https://doi.org/10.3390/v13061013








