Comparative Molecular Characterization of Novel and Known Piscine Toti-Like Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origin of Samples and Virus
2.2. RNA Extraction, Library Preparation, and NGS
2.2.1. GSTLV-1, CCTLV-1, and BGTLV-1
2.2.2. CLuTLV
2.3. Sanger Sequencing of CLuTLV
2.4. Phylogeny
2.5. In Silico Sequence Analyses on Full Length Genomic Sequences and Protein Sequence Derivatives
3. Results
3.1. Full-Length Genomic Sequence of Golden Shiner Toti-Like Virus 1 (GSTLV-1)
3.2. Novel Toti-Like Viruses Detected in Common Carp and Bluegill
3.3. Novel Toti-Like Virus Detected in Lumpsucker
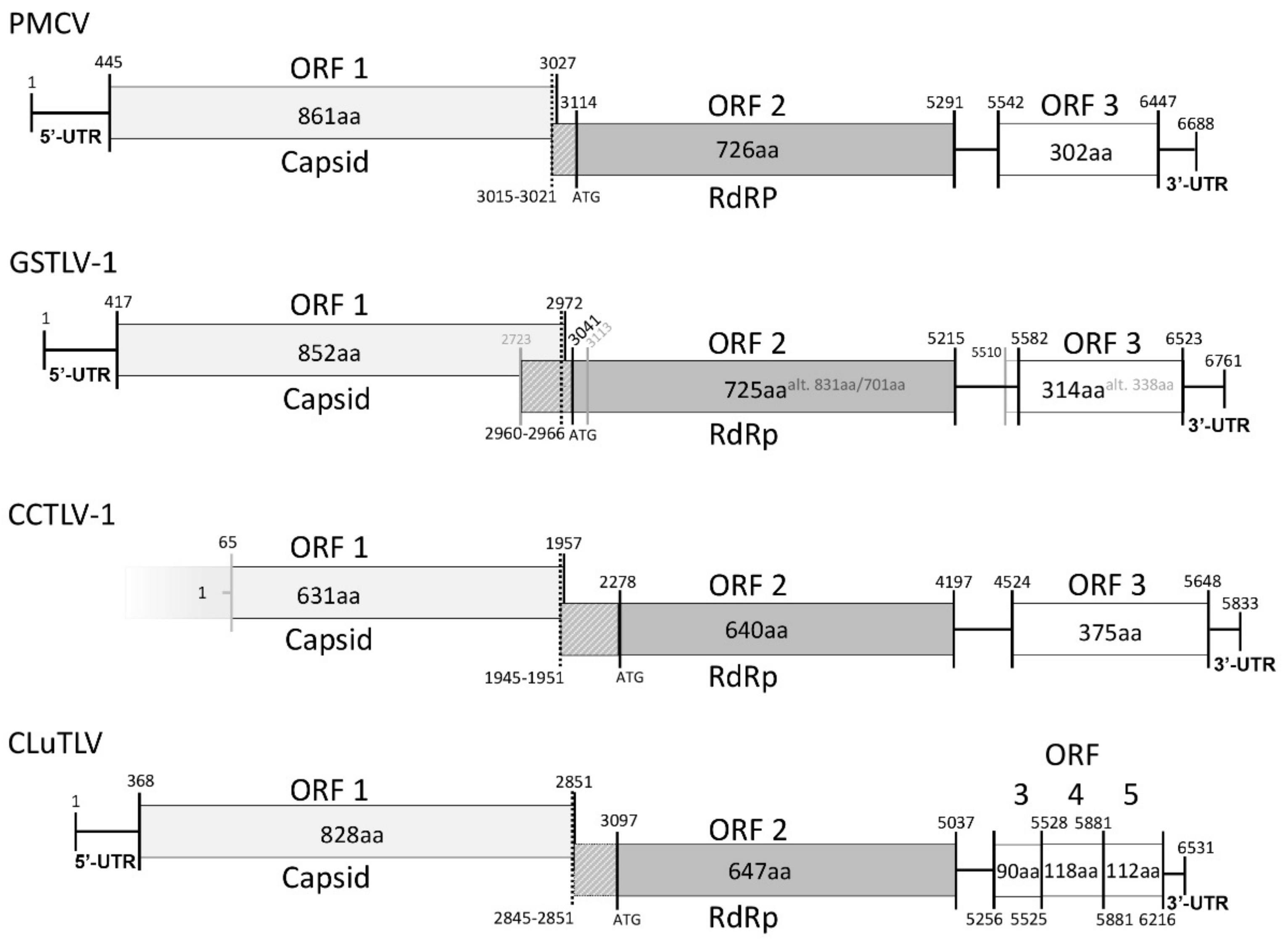
3.4. Piscine Toti-Like Viruses Share Major Genomic Characteristics
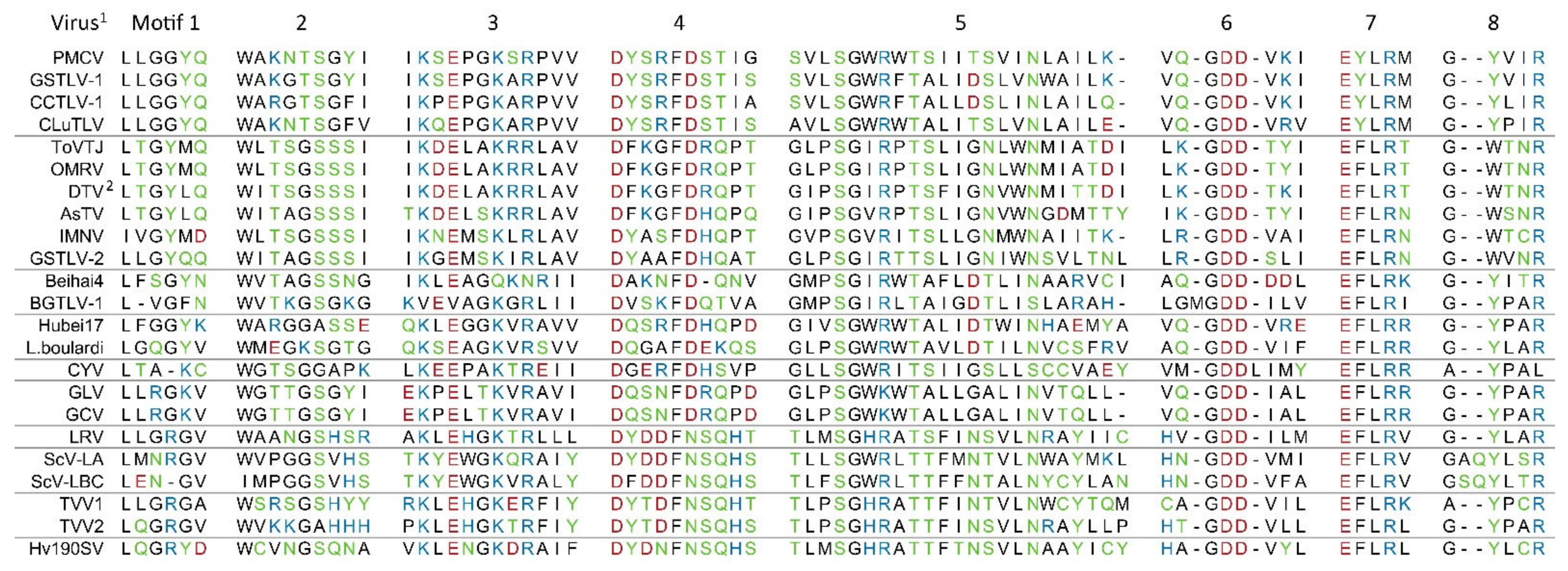
3.5. Characteristics of the Putative Capsid Encoded by ORF1 and RdRp Encoded by ORF2
3.6. Characteristics of ORF3 Encoded Protein in PMCV, GSTLV-1, and CCTLV-1
3.7. Characteristics of CLuTLV ORF 3, 4, and 5
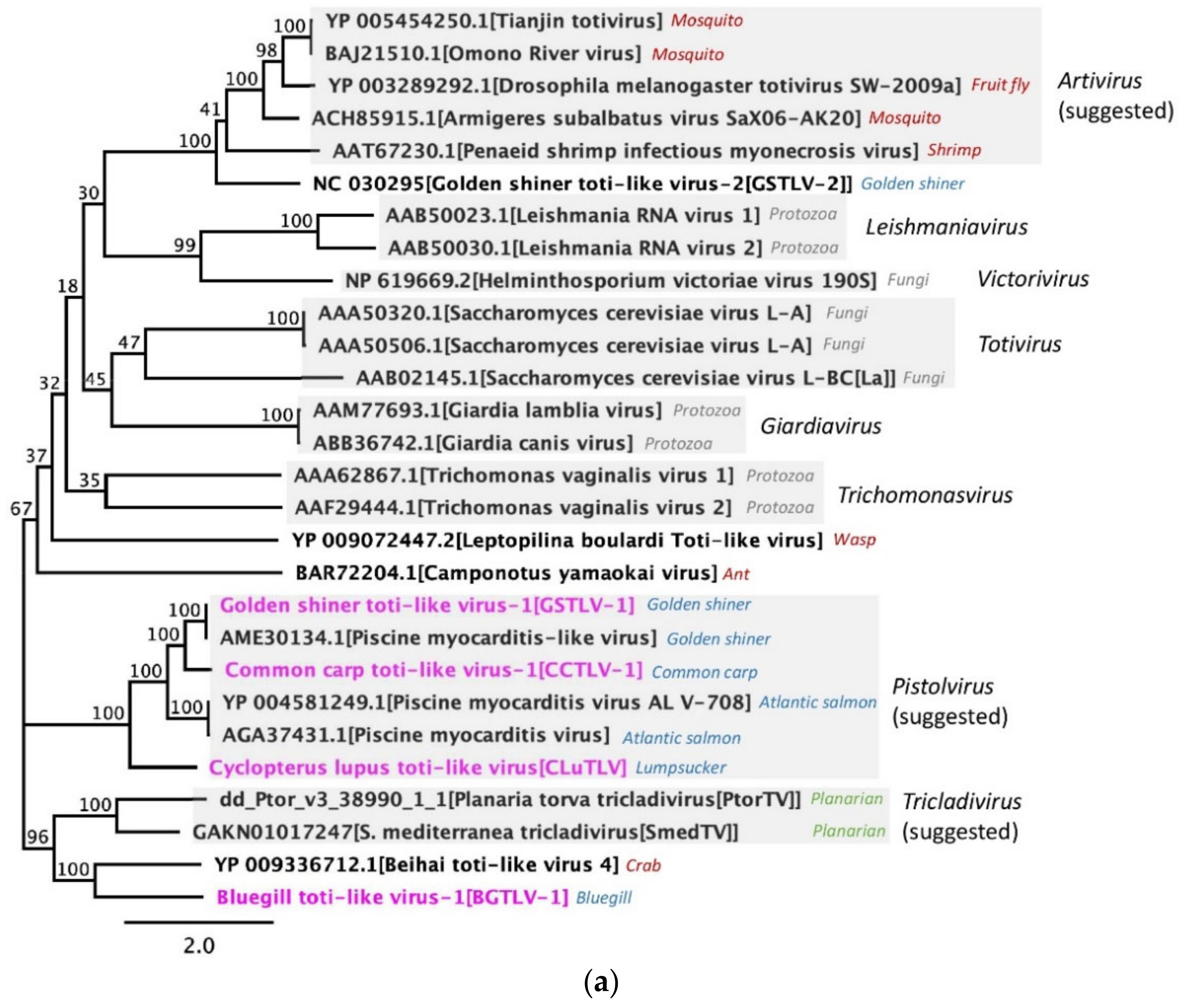
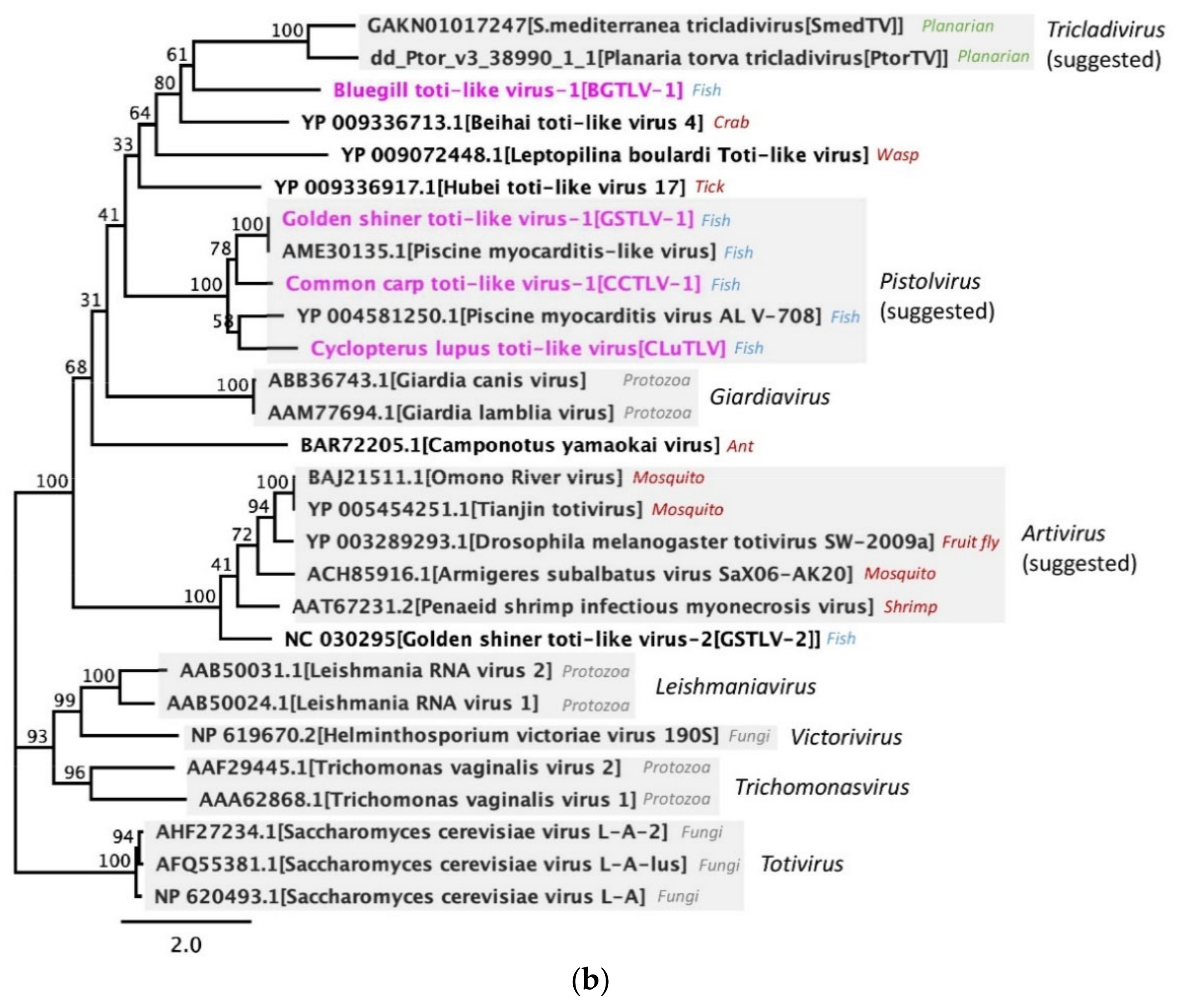
3.8. Phylogenetic Relation between Piscine Toti-Like Viruses and to Other Toti-Like Viruses and Totiviridae
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virus Taxonomy: The Classification and Nomenclature of Viruses—The Online (10th) Report of the ICTV, 10th ed.; International Committee on Taxonomy of Viruses (ICTV): London, UK, 2019.
- Isawa, H.; Kuwata, R.; Hoshino, K.; Tsuda, Y.; Sakai, K.; Watanabe, S.; Nishimura, M.; Satho, T.; Kataoka, M.; Nagata, N.; et al. Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res. 2011, 155, 147–155. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Attoui, H.; Mohd, J.F.; Wang, H.Q.; Cao, Y.X.; Fan, S.P.; Sun, Y.X.; Liu, L.D.; Mertens, P.P.; Meng, W.S.; et al. Isolation and full-length sequence analysis of Armigeres subalbatus totivirus, the first totivirus isolate from mosquitoes representing a proposed novel genus (Artivirus) of the family Totiviridae. J. Gen. Virol. 2010, 91, 2836–2845. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.P.; Shyam, K.U.; Banu, H.; Jeena, K.; Krishnan, R. Infectious Myonecrosis Virus (IMNV)—An alarming viral pathogen to Penaeid shrimps. Aquaculture 2017, 477, 99–105. [Google Scholar] [CrossRef]
- Burrows, J.T.A.; Depierreux, D.; Nibert, M.L.; Pearson, B.J. A Novel Taxon of Monosegmented Double-Stranded RNA Viruses Endemic to Triclad Flatworms. J. Virol. 2020, 94, e00623-20. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Ge, X.; Yuan, J.; Shi, Z. A novel totivirus-like virus isolated from bat guano. Arch. Virol. 2012, 157, 1093–1099. [Google Scholar] [CrossRef]
- Haugland, Ø.; Mikalsen, A.B.; Nilsen, P.; Lindmo, K.; Thu, B.J.; Eliassen, T.M.; Roos, N.; Rode, M.; Evensen, Ø. Cardiomyopathy syndrome of Atlantic salmon (Salmo salar L.) is caused by a dsRNA virus of the Totiviridae family. J. Virol. 2011, 85, 5275–5286. [Google Scholar] [CrossRef] [Green Version]
- Mor, S.K.; Phelps, N.B. Molecular detection of a novel totivirus from golden shiner (Notemigonus crysoleucas) baitfish in the USA. Arch. Virol. 2016, 161, 2227–2234. [Google Scholar] [CrossRef] [PubMed]
- Mor, S.K.; Phelps, N.B. Detection and molecular characterization of a novel piscine-myocarditis-like virus from baitfish in the USA. Arch. Virol. 2016, 161, 1925–1931. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.L.; Di Giallonardo, F.; Wille, M.; Ortiz-Baez, A.S.; Costa, V.A.; Ghaly, T.; Mifsud, J.C.O.; Turnbull, O.M.H.; Bellwood, D.R.; Williamson, J.E.; et al. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 2021, 7, veab005. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Di Cicco, E.; Günther, O.P.; Schulze, A.D.; Kaukinen, K.H.; Li, S.; Tabata, A.; Ming, T.J.; Ferguson, H.W.; Suttle, C.A.; et al. Discovery and surveillance of viruses from salmon in British Columbia using viral immune-response biomarkers, metatranscriptomics, and high-throughput RT-PCR. Virus Evol. 2021, 7, veaa069. [Google Scholar] [CrossRef]
- Poulos, B.T.; Tang, K.F.; Pantoja, C.R.; Bonami, J.R.; Lightner, D.V. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J. Gen. Virol. 2006, 87, 987–996. [Google Scholar] [CrossRef]
- Nibert, M.L.; Takagi, Y. Fibers come and go: Differences in cell-entry components among related dsRNA viruses. Curr. Opin. Virol. 2013, 3, 20–26. [Google Scholar] [CrossRef]
- Wiik-Nielsen, J.; Alarcon, M.; Fineid, B.; Rode, M.; Haugland, O. Genetic variation in Norwegian piscine myocarditis virus in Atlantic salmon, Salmo salar L. J. Fish Dis. 2012, 36, 11. [Google Scholar]
- Trimmomatic. Available online: https://github.com/usadellab/Trimmomatic (accessed on 2 June 2021).
- Bowtie2. Available online: https://github.com/BenLangmead/bowtie2 (accessed on 2 June 2021).
- SPAdes. Available online: https://github.com/ablab/spades (accessed on 2 June 2021).
- Zhang, K.Y.; Gao, Y.Z.; Du, M.Z.; Liu, S.; Dong, C.; Guo, F.B. Vgas: A Viral Genome Annotation System. Front. Microbiol. 2019, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.H.; Lu, C.L.; Chiu, H.T. A heuristic approach for detecting RNA H-type pseudoknots. Bioinformatics 2005, 21, 3501–3508. [Google Scholar] [CrossRef] [Green Version]
- Janssen, S.; Giegerich, R. The RNA shapes studio. Bioinformatics 2015, 31, 423–425. [Google Scholar] [CrossRef] [Green Version]
- KnotInFrame. Available online: https://bibiserv.cebitec.uni-bielefeld.de/knotinframe (accessed on 2 June 2021).
- Expasy compute pI/Mw. Available online: https://web.expasy.org/compute_pi/ (accessed on 2 June 2021).
- Hofman, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler 1993, 347, 166. [Google Scholar]
- TMpred. Available online: http://embnet.vital-it.ch/software/TMPRED_form.html (accessed on 2 June 2021).
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- SignalP 5.0. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 2 June 2021).
- PSORTII. Available online: http://www.genscript.com/psort.html (accessed on 2 June 2021).
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Ketteler, R. On programmed ribosomal frameshifting: The alternative proteomes. Front. Genet. 2012, 3, 242. [Google Scholar] [CrossRef] [Green Version]
- Plant, E.P.; Dinman, J.D. Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems. RNA 2006, 12, 666–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nibert, M.L. ‘2A-like’ and ‘shifty heptamer’ motifs in penaeid shrimp infectious myonecrosis virus, a monosegmented double-stranded RNA virus. J. Gen. Virol. 2007, 88, 1315–1318. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 1993, 21, 5667–5669. [Google Scholar] [CrossRef]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [CrossRef]
- Poimala, A.; Vainio, E.J. Complete genome sequence of a novel toti-like virus from the plant-pathogenic oomycete Phytophthora cactorum. Arch. Virol. 2020, 165, 1679–1682. [Google Scholar] [CrossRef] [PubMed]
- Shmulevitz, M.; Duncan, R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000, 19, 902–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas, M.D.; Cavalcante, G.H.; Oliveira, R.A.; Lanza, D.C. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res. 2016, 211, 159–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, S.; Urayama, S.; Ohmatsu, T.; Sassa, Y.; Sakai, C.; Takata, M.; Hayashi, S.; Nagai, M.; Furuya, T.; Moriyama, H.; et al. Identification, characterization and full-length sequence analysis of a novel dsRNA virus isolated from the arboreal ant Camponotus yamaokai. J. Gen. Virol. 2015, 96, 1930–1937. [Google Scholar] [CrossRef] [Green Version]
- Park, C.M.; Lopinski, J.D.; Masuda, J.; Tzeng, T.H.; Bruenn, J.A. A second double-stranded RNA virus from yeast. Virology 1996, 216, 451–454. [Google Scholar] [CrossRef]
- Goodman, R.P.; Freret, T.S.; Kula, T.; Geller, A.M.; Talkington, M.W.; Tang-Fernandez, V.; Suciu, O.; Demidenko, A.A.; Ghabrial, S.A.; Beach, D.H.; et al. Clinical isolates of Trichomonas vaginalis concurrently infected by strains of up to four Trichomonasvirus species (Family Totiviridae). J. Virol. 2011, 85, 4258–4270. [Google Scholar] [CrossRef] [Green Version]
- Amono, R.; Evensen, Ø.; Mikalsen, A.B. Characteristics of PMCV ORF3 post expression peptides. Unpublished work. 2021. [Google Scholar]
- Boutilier, J.; Duncan, R. The reovirus fusion-associated small transmembrane (FAST) proteins: Virus-encoded cellular fusogens. Curr. Top Membr. 2011, 68, 107–140. [Google Scholar]
- Lovoll, M.; Alarcon, M.; Bang, J.B.; Taksdal, T.; Kristoffersen, A.B.; Tengs, T. Quantification of piscine reovirus (PRV) at different stages of Atlantic salmon Salmo salar production. Dis. Aquat. Organ. 2012, 99, 7–12. [Google Scholar] [CrossRef] [Green Version]
- Key, T.; Read, J.; Nibert, M.L.; Duncan, R. Piscine reovirus encodes a cytotoxic, non-fusogenic, integral membrane protein and previously unrecognized virion outer-capsid proteins. J. Gen. Virol. 2013, 94, 1039–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson, H.W.; Poppe, T.; Speare, D.J. Cardiomyopathy in farmed Norwegian salmon. Dis. Aquat. Organ. 1990, 8, 225–231. [Google Scholar] [CrossRef]
- Fritsvold, C.; Kongtorp, R.T.; Taksdal, T.; Orpetveit, I.; Heum, M.; Poppe, T.T. Experimental transmission of cardiomyopathy syndrome (CMS) in Atlantic salmon Salmo salar. Dis. Aquat. Organ. 2009, 87, 225–234. [Google Scholar] [CrossRef]
- Rodger, H.; Turnbull, T. Cardiomyopathy syndrome in farmed Scottish salmon. Vet. Rec. 2000, 146, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Rodger, H.D.; McCleary, S.J.; Ruane, N.M. Clinical cardiomyopathy syndrome in Atlantic salmon, Salmo salar L. J. Fish. Dis. 2014, 37, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Pantoja, C.R.; Poulos, B.T.; Redman, R.M.; Lightnere, D.V. In situ hybridization demonstrates that Litopenaeus vannamei L. stylirostris and Penaeus monodon are susceptible to experimental infection with infectious myonecrosis virus (IMNV). Dis. Aquat. Organ. 2005, 63, 261–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| (a) | ||||||||
| PMCV | GSTLV-1 | CCTLV-1 | CLuTLV | BGTLV-1 | GSTLV-2 | |||
| Total nt | 6688 | 6761 | >5833 2 | 6351 | >5307 2,3 | 7788 3 | ||
| 5′ UTR nt | 444 | 416 | >64 2 | 367 | >192 2 | 135 | ||
| 3′ UTR nt | 241 | 235 | >185 2 | 132 | >97 3 | >54 3 | ||
| ORF 1/Capsid | ||||||||
| Peptides aa | n.a 4 | n.a 4 | n.a 4 | n.a 4 | n.a 4 | 312 161 319 | ||
| Capsid aa | 861 | 852 | >631 2 | 828 | 891 | 867 3 | ||
| Capsid kDa | 91.8 | 92.0 | n.a. 2,4 | 90.5 | n.a. 2,4 | 96.3 | ||
| pI of capsid | 5.4 | 5.8 | n.a. 2,4 | 5.0 | n.a. 2,4 | 5.47 | ||
| GC (%) | 56.2 | 51.4 | n.a. 2,4 | 49.6 | n.a. 2,4 | 37.2 | ||
| ORF2/RdRp | ||||||||
| aa | 726 | 725 | 640 | 647 | 785 | 737 | ||
| kDa | 83.1 | 94.8 | 73.5 | 74.2 | 88.9 | 85.8 | ||
| pI | 9.4 | 9.4 | 9.1 | 9.3 | 8.6 | 9.5 | ||
| GC (%) | 43.6 | 40.8 | 39.7 | 41.8 | 48.5 | 27.2 | ||
| ORF3 | ORF3 | ORF4 | ORF5 | |||||
| aa | 302 | 314 (338) 1 | 375 | 90 | 118 | 112 | n.a 4 | n.a 4 |
| kDa | 33.4 | 34.5 (37.3) 1 | 42.2 | 9.7 | 14.0 | 13.2 | n.a 4 | n.a 4 |
| pI | 7.0 | 9.2 (9.3) 1 | 6.5 | 5.2 | 6.1 | 4.4 | n.a 4 | n.a 4 |
| GC (%) | 45.0 | 42.1 (41.8) 1 | 44.3 | 48.5 | 46.3 | 39.9 | n.a 4 | n.a 4 |
| (b) | ||||||||
| PMCV | GSTLV-1 | CCTLV-1 | CLuTLV | BGTLV-1 | GSTLV-2 | PMCV | GSTLV-1 | |
| Capsid-RdRp | ||||||||
| aa | 1616 | 1600 | >1378 2 | 1557 | 1673 | 1740 | 1616 | 1600 |
| kDa | 178.2 | 177.0 | n.a. 2,4 | 174.2 | n.a. 2,4 | 198.2 | 178.2 | 177.0 |
| pI | 8.9 | 8.0 | n.a. 2,4 | 6.3 | n.a. 2,4 | 8.8 | 8.9 | 8.0 |
| RdRp-part of fused | ||||||||
| aa | 757 | 750 | 749 | 729 | 782–785 5 | 903 | 757 | 750 |
| kDa | 86.6 | 85.3 | 86.0 | 83.7 | 88.6–88.9 | 105.2 | 86.6 | 85.3 |
| pI | 9.6 | 9.0 | 8.8 | 9.3 | 8.6 | 9.3 | 9.6 | 9.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandlund, L.; Mor, S.K.; Singh, V.K.; Padhi, S.K.; Phelps, N.B.D.; Nylund, S.; Mikalsen, A.B. Comparative Molecular Characterization of Novel and Known Piscine Toti-Like Viruses. Viruses 2021, 13, 1063. https://doi.org/10.3390/v13061063
Sandlund L, Mor SK, Singh VK, Padhi SK, Phelps NBD, Nylund S, Mikalsen AB. Comparative Molecular Characterization of Novel and Known Piscine Toti-Like Viruses. Viruses. 2021; 13(6):1063. https://doi.org/10.3390/v13061063
Chicago/Turabian StyleSandlund, Liv, Sunil K. Mor, Vikash K. Singh, Soumesh K. Padhi, Nicholas B. D. Phelps, Stian Nylund, and Aase B. Mikalsen. 2021. "Comparative Molecular Characterization of Novel and Known Piscine Toti-Like Viruses" Viruses 13, no. 6: 1063. https://doi.org/10.3390/v13061063
APA StyleSandlund, L., Mor, S. K., Singh, V. K., Padhi, S. K., Phelps, N. B. D., Nylund, S., & Mikalsen, A. B. (2021). Comparative Molecular Characterization of Novel and Known Piscine Toti-Like Viruses. Viruses, 13(6), 1063. https://doi.org/10.3390/v13061063







