The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation
Abstract
:1. Introduction
2. The Biological Function of the SARS-CoV-2 N Protein
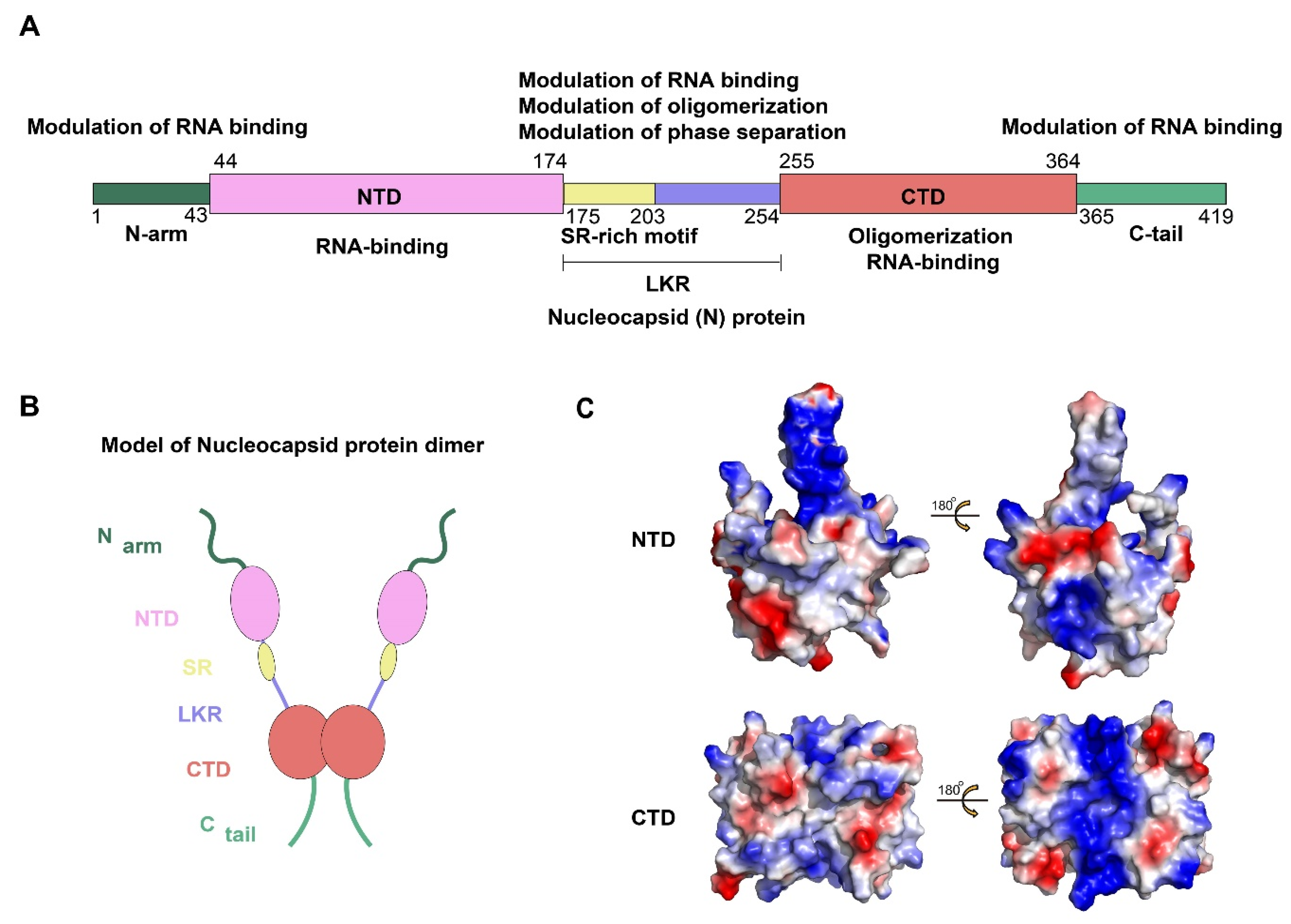
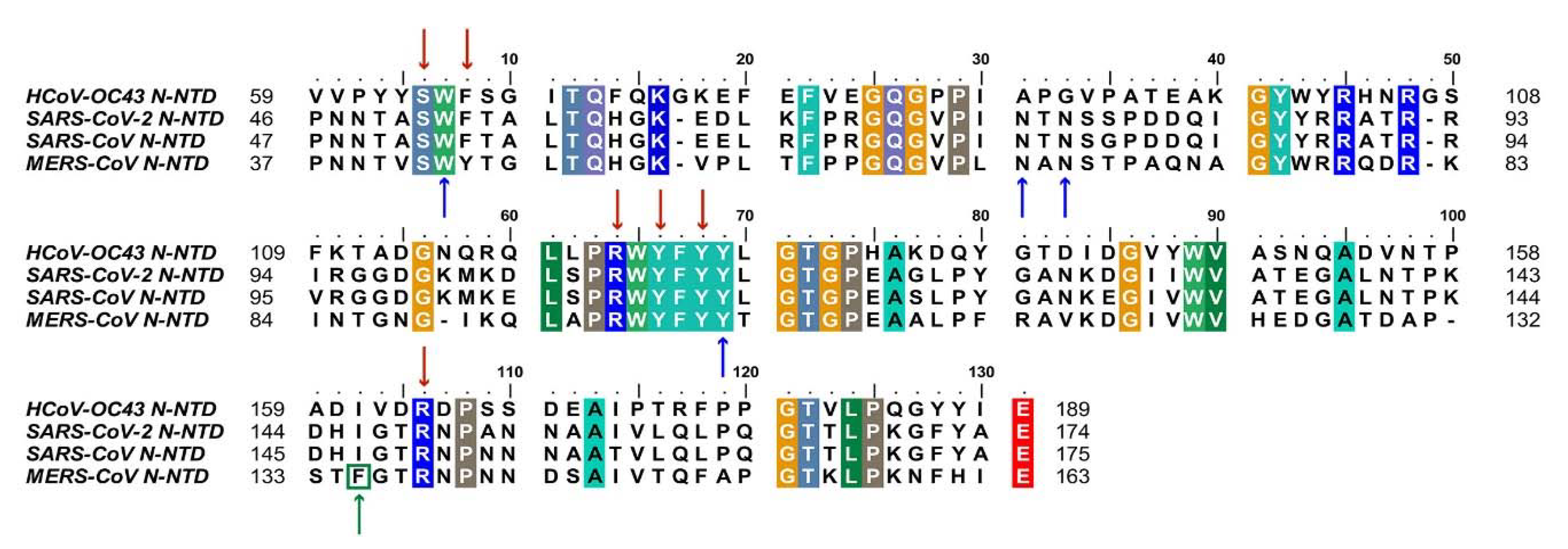
2.1. Composition and Structure of SARS-CoV-2 N Protein
2.2. Genome Encapsidation: The Primary Function of a Viral N Protein
2.3. N Protein Undergoes Liquid-Liquid Phase Separation to Enhance Viral Transcription and Assembly
2.4. Regulation of the Host’ Immune Response
3. Clinical Applications of the SARS CoV-2 N Protein
3.1. N-Protein as a Diagnostic Marker
3.2. N Protein: As a Therapeutic Target
4. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Papatsiros, V.G.; Stylianaki, I.; Papakonstantinou, G.; Papaioannou, N.; Christodoulopoulos, G. Case Report of Transmissible Gastroenteritis Coronavirus Infection Associated with Small Intestine and Brain Lesions in Piglets. Viral Immunol. 2019, 32, 63–67. [Google Scholar] [CrossRef]
- Cavanagh, D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zhou, Y.; Wu, H.; Kou, Z.; Liu, S.; Jiang, S. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004, 42, 5309–5314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, P.S.; Sturman, L.S. Background Paper Functions of the Coronavirus Nucleocapsid Protein. In Coronaviruses and Their Diseases; Springer: Berlin/Heidelberg, Germany, 1990; pp. 235–238. [Google Scholar]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surjit, M.; Liu, B.; Chow, V.T.; Lal, S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006, 281, 10669–10681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Li, J.; Liu, X.E.; Wang, L.; Li, T.; Zhou, Y.H.; Zhuang, H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: Comparison with results of other viral markers. J. Virol. Methods 2005, 130, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.K.; Sue, S.C.; Yu, T.H.; Hsieh, C.M.; Tsai, C.K.; Chiang, Y.C.; Lee, S.J.; Hsiao, H.H.; Wu, W.J.; Chang, W.L.; et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006, 13, 59–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, D.C.; Chalupska, D.; Silhan, J.; Koutna, E.; Nencka, R.; Veverka, V.; Boura, E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020, 16, e1009100. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Chen, J.; Chen, K.; Shen, X.; Jiang, H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: Self-association analysis and nucleic acid binding characterization. Biochemistry 2006, 45, 11827–11835. [Google Scholar] [CrossRef]
- Chang, C.K.; Hsu, Y.L.; Chang, Y.H.; Chao, F.A.; Wu, M.C.; Huang, Y.S.; Hu, C.K.; Huang, T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: Implications for ribonucleocapsid protein packaging. J. Virol. 2009, 83, 2255–2264. [Google Scholar] [CrossRef] [Green Version]
- Giri, R.; Bhardwaj, T.; Shegane, M.; Gehi, B.R.; Kumar, P.; Gadhave, K.; Oldfield, C.J.; Uversky, V.N. Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. Cell. Mol. Life Sci. 2021, 78, 1655–1688. [Google Scholar] [CrossRef]
- Schiavina, M.; Pontoriero, L.; Uversky, V.N.; Felli, I.C.; Pierattelli, R. The highly flexible disordered regions of the SARS-CoV-2 nucleocapsid N protein within the 1-248 residue construct: Sequence-specific resonance assignments through NMR. Biomol. NMR Assign. 2021, 15, 219–227. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chang, C.K.; Chang, Y.W.; Sue, S.C.; Bai, H.I.; Riang, L.; Hsiao, C.D.; Huang, T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007, 368, 1075–1086. [Google Scholar] [CrossRef]
- Takeda, M.; Chang, C.K.; Ikeya, T.; Güntert, P.; Chang, Y.H.; Hsu, Y.L.; Huang, T.H.; Kainosho, M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 2008, 380, 608–622. [Google Scholar] [CrossRef]
- Peng, T.Y.; Lee, K.R.; Tarn, W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008, 275, 4152–4163. [Google Scholar] [CrossRef] [Green Version]
- Hilser, V.J.; Thompson, E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 8311–8315. [Google Scholar] [CrossRef] [Green Version]
- Surjit, M.; Liu, B.; Kumar, P.; Chow, V.T.; Lal, S.K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004, 317, 1030–1036. [Google Scholar] [CrossRef]
- He, R.; Dobie, F.; Ballantine, M.; Leeson, A.; Li, Y.; Bastien, N.; Cutts, T.; Andonov, A.; Cao, J.; Booth, T.F.; et al. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004, 316, 476–483. [Google Scholar] [CrossRef]
- Yu, I.M.; Gustafson, C.L.; Diao, J.; Burgner, J.W., 2nd; Li, Z.; Zhang, J.; Chen, J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005, 280, 23280–23286. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; West, A.M.V.; Silletti, S.; Corbett, K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci 2020, 29, 1890–1901. [Google Scholar] [CrossRef]
- Surjit, M.; Kumar, R.; Mishra, R.N.; Reddy, M.K.; Chow, V.T.; Lal, S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol 2005, 79, 11476–11486. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.K.; Chen, C.M.; Chiang, M.H.; Hsu, Y.L.; Huang, T.H. Transient oligomerization of the SARS-CoV N protein--implication for virus ribonucleoprotein packaging. PLoS ONE 2013, 8, e65045. [Google Scholar] [CrossRef] [PubMed]
- Novoa, R.R.; Calderita, G.; Arranz, R.; Fontana, J.; Granzow, H.; Risco, C. Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol. Cell 2005, 97, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Holehouse, A.S.; Pappu, R.V. Functional Implications of Intracellular Phase Transitions. Biochemistry 2018, 57, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Savastano, A.; Ibáñez de Opakua, A.; Rankovic, M.; Zweckstetter, M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020, 11, 6041. [Google Scholar] [CrossRef]
- Carlson, C.R.; Asfaha, J.B.; Ghent, C.M.; Howard, C.J.; Hartooni, N.; Safari, M.; Frankel, A.D.; Morgan, D.O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Mol. Cell 2020, 80, 1092–1103.e1094. [Google Scholar] [CrossRef]
- Iserman, C.; Roden, C.A.; Boerneke, M.A.; Sealfon, R.S.G.; McLaughlin, G.A.; Jungreis, I.; Fritch, E.J.; Hou, Y.J.; Ekena, J.; Weidmann, C.A.; et al. Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol. Cell 2020, 80, 1078–1091.e1076. [Google Scholar] [CrossRef]
- Chen, H.; Cui, Y.; Han, X.; Hu, W.; Sun, M.; Zhang, Y.; Wang, P.H.; Song, G.; Chen, W.; Lou, J. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. 2020, 30, 1143–1145. [Google Scholar] [CrossRef] [PubMed]
- Perdikari, T.M.; Murthy, A.C.; Ryan, V.H.; Watters, S.; Naik, M.T.; Fawzi, N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020, 39, e106478. [Google Scholar] [CrossRef]
- Cascarina, S.M.; Ross, E.D. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020, 34, 9832–9842. [Google Scholar] [CrossRef]
- Guseva, S.; Milles, S.; Jensen, M.R.; Salvi, N.; Kleman, J.P.; Maurin, D.; Ruigrok, R.W.H.; Blackledge, M. Measles virus nucleo-and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci. Adv. 2020, 6, eaaz7095. [Google Scholar] [CrossRef] [Green Version]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Shibabaw, T.; Molla, M.D.; Teferi, B.; Ayelign, B. Role of IFN and Complements System: Innate Immunity in SARS-CoV-2. J. Inflamm. Res. 2020, 13, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Amor, S.; Fernández Blanco, L.; Baker, D. Innate immunity during SARS-CoV-2: Evasion strategies and activation trigger hypoxia and vascular damage. Clin. Exp. Immunol. 2020, 202, 193–209. [Google Scholar] [CrossRef]
- Ding, S.W.; Han, Q.; Wang, J.; Li, W.X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 2018, 54, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [Green Version]
- Mu, J.; Xu, J.; Zhang, L.; Shu, T.; Wu, D.; Huang, M.; Ren, Y.; Li, X.; Geng, Q.; Xu, Y.; et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020, 63, 1413–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Sastre, A.; Biron, C.A. Type 1 interferons and the virus-host relationship: A lesson in détente. Science 2006, 312, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 84. [Google Scholar] [CrossRef]
- Li, J.Y.; Liao, C.H.; Wang, Q.; Tan, Y.J.; Luo, R.; Qiu, Y.; Ge, X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhuang, M.W.; Han, L.; Zhang, J.; Nan, M.L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020, 5, 299. [Google Scholar] [CrossRef]
- Chen, K.; Xiao, F.; Hu, D.; Ge, W.; Tian, M.; Wang, W.; Pan, P.; Wu, K.; Wu, J. SARS-CoV-2 Nucleocapsid Protein Interacts with RIG-I and Represses RIG-Mediated IFN-β Production. Viruses 2021, 13, 47. [Google Scholar] [CrossRef]
- Mu, J.; Fang, Y.; Yang, Q.; Shu, T.; Wang, A.; Huang, M.; Jin, L.; Deng, F.; Qiu, Y.; Zhou, X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020, 6, 65. [Google Scholar] [CrossRef]
- Asselah, T.; Durantel, D.; Pasmant, E.; Lau, G.; Schinazi, R.F. COVID-19: Discovery, diagnostics and drug development. J. Hepatol. 2021, 74, 168–184. [Google Scholar] [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef]
- Meyer, B.; Drosten, C.; Müller, M.A. Serological assays for emerging coronaviruses: Challenges and pitfalls. Virus Res. 2014, 194, 175–183. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Qu, J.; Wu, C.; Li, X.; Zhang, G.; Jiang, Z.; Li, X.; Zhu, Q.; Liu, L. Profile of Immunoglobulin G and IgM Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 2255–2258. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg. Infect. Dis. 2020, 26, 1478–1488. [Google Scholar] [CrossRef]
- Li, T.; Wang, L.; Wang, H.; Li, X.; Zhang, S.; Xu, Y.; Wei, W. Serum SARS-COV-2 Nucleocapsid Protein: A Sensitivity and Specificity Early Diagnostic Marker for SARS-COV-2 Infection. Front. Cell. Infect. Microbiol. 2020, 10, 470. [Google Scholar] [CrossRef] [PubMed]
- Amrun, S.N.; Lee, C.Y.; Lee, B.; Fong, S.W.; Young, B.E.; Chee, R.S.; Yeo, N.K.; Torres-Ruesta, A.; Carissimo, G.; Poh, C.M.; et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 2020, 58, 102911. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.Y.; Liao, G.R.; Lien, A.; Hsu, W.L. Current concepts in the development of therapeutics against human and animal coronavirus diseases by targeting NP. Comput. Struct. Biotechnol. J. 2021, 19, 1072–1080. [Google Scholar] [CrossRef]
- Chang, C.K.; Jeyachandran, S.; Hu, N.J.; Liu, C.L.; Lin, S.Y.; Wang, Y.S.; Chang, Y.M.; Hou, M.H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. Biosyst. 2016, 12, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.M.; Lin, S.C.; Hsu, J.N.; Chang, C.K.; Chien, C.M.; Wang, Y.S.; Wu, H.Y.; Jeng, U.S.; Kehn-Hall, K.; Hou, M.H. Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design. J. Med. Chem. 2020, 63, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Hayouka, Z.; Rosenbluh, J.; Levin, A.; Loya, S.; Lebendiker, M.; Veprintsev, D.; Kotler, M.; Hizi, A.; Loyter, A.; Friedler, A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. USA 2007, 104, 8316–8321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.F.; Chen, Y.H.; Chu, S.Y.; Lin, M.I.; Hsu, H.T.; Wu, P.Y.; Wu, C.J.; Liu, H.W.; Lin, F.Y.; Lin, G.; et al. E339...R416 salt bridge of nucleoprotein as a feasible target for influenza virus inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 16515–16520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, Y.S.; Lin, S.Y.; Wang, S.M.; Wang, C.T.; Chiu, Y.L.; Huang, T.H.; Hou, M.H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013, 587, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Roh, C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed. 2012, 7, 2173–2179. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.; Sridhar, S.; Chiu, K.H.; Hung, D.L.; Li, X.; Hung, I.F.; Tam, A.R.; Chung, T.W.; Chan, J.F.; Zhang, A.J.; et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg. Microbes Infect. 2021, 10, 507–535. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Wang, M.X.; Ang, I.Y.H.; Tan, S.H.X.; Lewis, R.F.; Chen, J.I.; Gutierrez, R.A.; Gwee, S.X.W.; Chua, P.E.Y.; Yang, Q.; et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J. Clin. Med. 2020, 9, 623. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.H.; Strych, U.; Hotez, P.J.; Bottazzi, M.E. The SARS-CoV-2 Vaccine Pipeline: An Overview. Curr. Trop. Med. Rep. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ruan, Y.J.; Wei, C.L.; Ee, A.L.; Vega, V.B.; Thoreau, H.; Su, S.T.; Chia, J.M.; Ng, P.; Chiu, K.P.; Lim, L.; et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 2003, 361, 1779–1785. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.Y.; Werner, H.C.; Kong, W.P.; Leung, K.; Traggiai, E.; Lanzavecchia, A.; Nabel, G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. USA 2005, 102, 797–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Ulasli, M.; Schepers, H.; Mauthe, M.; V’Kovski, P.; Kriegenburg, F.; Thiel, V.; de Haan, C.A.M.; Reggiori, F. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, D.T.; Tam, F.C.; Ma, C.H.; Chan, P.K.; Cheung, J.L.; Niu, H.; Tam, J.S.; Lim, P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004, 190, 379–386. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Tamin, A.; Soloff, A.; D’Aiuto, L.; Nwanegbo, E.; Robbins, P.D.; Bellini, W.J.; Barratt-Boyes, S.; Gambotto, A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet 2003, 362, 1895–1896. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Takemoto, Y.; Okuno, Y.; Hashimoto, S.; Yoshida, S.; Fukunaga, Y.; Tanaka, T.; Kita, Y.; Kuwayama, S.; Muraki, Y.; et al. The development of vaccines against SARS corona virus in mice and SCID-PBL/hu mice. Vaccine 2005, 23, 2269–2272. [Google Scholar] [CrossRef]
- Zhou, R.; To, K.K.; Wong, Y.C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.Y.; Lau, T.T.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e865. [Google Scholar] [CrossRef]
- Cao, W.C.; Liu, W.; Zhang, P.H.; Zhang, F.; Richardus, J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007, 357, 1162–1163. [Google Scholar] [CrossRef]
- Ng, O.W.; Chia, A.; Tan, A.T.; Jadi, R.S.; Leong, H.N.; Bertoletti, A.; Tan, Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine 2016, 34, 2008–2014. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Oliveira, S.C.; de Magalhães, M.T.Q.; Homan, E.J. Immunoinformatic Analysis of SARS-CoV-2 Nucleocapsid Protein and Identification of COVID-19 Vaccine Targets. Front. Immunol. 2020, 11, 587615. [Google Scholar] [CrossRef]
- Ahlén, G.; Frelin, L.; Nikouyan, N.; Weber, F.; Höglund, U.; Larsson, O.; Westman, M.; Tuvesson, O.; Gidlund, E.K.; Cadossi, M.; et al. The SARS-CoV-2 N Protein Is a Good Component in a Vaccine. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Liu, C.L.; Chang, Y.M.; Zhao, J.; Perlman, S.; Hou, M.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014, 57, 2247–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iserman, C.; Roden, C.; Boerneke, M.; Sealfon, R.; McLaughlin, G.; Jungreis, I.; Park, C.; Boppana, A.; Fritch, E.; Hou, Y.J.; et al. Specific viral RNA drives the SARS CoV-2 nucleocapsid to phase separate. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wheeler, R.J.; Lee, H.O.; Poser, I.; Pal, A.; Doeleman, T.; Kishigami, S.; Kour, S.; Anderson, E.N.; Marrone, L.; Murthy, A.C.; et al. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]

| Compounds | Target Domain or Process | Mechanism | Reference |
|---|---|---|---|
| PJ34, N-(6-oxo-5,6-dihydrophenanthridin-2-yl) (N,N-dimethylamino) acetamide hydrochloride | NTD | Reduce RNA binding | [14,67] |
| H3, 6-chloro-7-(2-morpholin-4-ylethylamino) quinoxaline-5,8-dione | NTD | Reduce RNA binding | [61] |
| (−)-catechin gallate | NTD | Reduce RNA binding | [66] |
| (−)-gallocatechin gallate | NTD | Reduce RNA binding | [66] |
| P3, 5-benzyloxygr- amine | CTD | Induce abnormal dimerization | [62] |
| 1,6-hexanediol | LLPS | prevent condensate formation | [68] |
| Lipoic acid | LLPS | Reduce smaller condensate | [68,69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 2021, 13, 1115. https://doi.org/10.3390/v13061115
Bai Z, Cao Y, Liu W, Li J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses. 2021; 13(6):1115. https://doi.org/10.3390/v13061115
Chicago/Turabian StyleBai, Zhihua, Ying Cao, Wenjun Liu, and Jing Li. 2021. "The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation" Viruses 13, no. 6: 1115. https://doi.org/10.3390/v13061115
APA StyleBai, Z., Cao, Y., Liu, W., & Li, J. (2021). The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses, 13(6), 1115. https://doi.org/10.3390/v13061115






