Abstract
Increasing evidence suggests that some newly emerged SARS-CoV-2 variants of concern (VoCs) resist neutralization by antibodies elicited by the early-pandemic wild-type virus. We applied neutralization tests to paired recoveree sera (n = 38) using clinical isolates representing the first wave (D614G), VoC1, and VoC2 lineages (B.1.1.7 and B 1.351). Neutralizing antibodies inhibited contemporary and VoC1 lineages, whereas inhibition of VoC2 was reduced 8-fold, with 50% of sera failing to show neutralization. These results provide evidence for the increased potential of VoC2 to reinfect previously SARS-CoV-infected individuals. The kinetics of NAbs in different patients showed similar decline against all variants, with generally low initial anti-B.1.351 responses becoming undetectable, but with anti-B.1.1.7 NAbs remaining detectable (>20) for months after acute infection.
1. Introduction
Neutralizing antibodies (NAbs), most of which target the receptor binding domain (RBD) of viral spike protein, protect against SARS-CoV-2 infection [1,2,3,4,5,6]. However, some of the recently emerged variant SARS-CoV-2 lineages have raised concerns with regard to lowered protective immunity in vaccinees and recoverees due to mutations that cause neutralization escape [7,8,9,10]. Studies have indicated that the B.1.1.7-variant is neutralized almost equally to the original virus type, whereas increasing evidence suggests that the B.1.351 variant is less well neutralized.
Currently, no “golden standard” methodology exists for measuring NAbs against SARS-CoV-2, and assays have been performed with a variety of techniques such as SARS-CoV-2 spike-variant pseudotyped lentivirus [11,12,13], VSV [14,15,16], infectious clones, and virus isolates [17,18]. Cell lines have also varied, with some expressing transmembrane protease serine 2 (TMPRSS2) and/or angiotensin-converting enzyme 2 (ACE2), known to mediate SARS-CoV-2 entry. SARS-CoV-2 is known to readily adapt to Vero E6 (VE6) cells that have low TMPRSS2 but high ACE2 expression [19]. This can result in deletions around the furin cleavage site and force the viral entry to occur mainly via the endosomal route aided by alternate proteases.
In order to estimate to which extent and for how long antibodies raised by earlier infection protect against new variants of concern (VoC), and for optimizing the routine microneutralization tests (MNT) in use, we tested a panel of 38 paired sera from 18 laboratory-confirmed COVID-19 patients from spring and summer 2020 in MNT with different clinical isolates of SARS-CoV-2 including the B.1.1.7 and B 1.351 strains (now prevalent in the country) using Vero E6 (VE6) cells with and without expression of TMPRSS2.
2. Materials and Methods
2.1. Patient Samples
The samples included 38 sera from 18 laboratory-confirmed patients from spring and summer 2020 drawn 2–4 weeks after the disease and 2–8 months later (Table S1). Patients had either been recovering at home or were treated in the hospital in regular wards or in intensive care units (ICUs). The study was performed according to research and ethical permit of HUS (detailed in Institutional Review Board Statement), and informed consent was obtained from all patients.
2.2. Cell Lines
Vero E6 cells (VE6) and their TMRPRSS2-expressing clone VE6-TMPRSS2-H10 [20] (VE6T) were grown in minimal essential eagle’s medium (MEM, Sigma-Aldrich, Saint Louis, USA) including 10% (cell maintenance) or 2% (infection experiments) fetal bovine serum (FBS, ThermoFisher, Waltham, USA), 2 mM L-glutamine, 100 IU/mL penicillin, and 100 µg/mL streptomycin.
2.3. Virus Strains
Tests were performed with 4 clinical virus isolates: SARS-CoV-2/Finland/1/2020 (FIN-1), C1P1, VoC1, and VoC2. FIN-1 (passage 7) is a VE6 passaged virus strain that was isolated from the first patient in Finland in January 2020 [21]. C1P1 is a wild-type low-passage strain representing strains circulating in Finland during spring 2020 (with e.g., the D614G mutation), devoid of mutations around the furin-cleavage site [19]. Strains representing typical B.1.1.7 (VoC1) and B.1.351 (VoC2) strains (Table S2) were isolated in VE6T cells from patient nasopharyngeal samples (200 µL) as previously described [20] and used as low-passage (p1 and p0) stocks. Virus replication was determined by RdRp-targeting RT-PCR [22]. The infectious virus titers were determined by plaque assay in VE6T cells, and the isolates were sequenced as previously described (Table S2) [19].
2.4. Comparing Microneutralization Tests with Different Cell Lines and Virus Strains
Microneutralization tests were performed in a BSL3 level laboratory following the protocol published by Haveri et al. [21]. To address the host cell line effects with regard to neutralization, patient NAb titers from 1:20 serum dilution onwards were first compared in triplicate reactions both in VE6 cells and VE6T cells with the FIN-1 strain. VE6-TMPRSS2-H10 cells were also tested with C1P1-strain. Next, NAb titers against wild-type C1P1 and variant stains VoC1 and VoC2 were determined and compared using VE6T cells.
2.5. Determining Anti-NP- and Anti-Spike-Titers
Anti-Spike-IgG and anti-nucleoprotein-IgG titers were determined by ELISA against respective antigens based on the first strains reported from Wuhan in January 2020, as described previously [17,23,24].
2.6. Statistical Testing
Statistical tests were performed with IBM SPSS Statistics 25. Titers of <20 were set to 10 for calculations. Values 1, 5, and LOD/ were also tested for titers <20, but those did not change the significance levels (0.05, 0.01, and 0.001) of statistical tests between virus strains. Due to the data not being normally distributed, non-parametric related-samples Wilcoxon signed rank test and related-samples Friedman’s two-way analysis of variance by ranks tests were used for testing the significances of the differences between virus strains, and the non-parametric Mann–Whitney U test was used to test the differences between subgroups (0–150 days and over 150 days from the onset of symptoms, and whether the patient was treated at home or at hospital). To test the differences in NAb titer decrease rate between C1P1, VoC1, and VoC2, slopes of fit lines between paired samples of individual patients were calculated and compared with Friedman’s two-way analysis of variance by ranks test. Log2 transformed titers were used in the y-axis and days from the onset of the disease in the x-axis. Only samples with the determinable titer were used in this analysis.
3. Results
3.1. Optimizing MNT Protocol
We first wanted to ensure that the titers obtained reflected neutralization against circulating wild-type strains in cells with relevant entry molecules. Hence, we compared neutralizing antibody titers against FIN-1, a Wuhan-like strain from January 2020 using VE6 cells both with and without TMPRSS2 expression to a D614G isolate C1P1 using VE6 cells expressing TMPRSS2 (VE6T). The NAb titers in a microneutralization test (MNT) were significantly higher with the non-VE6-adapted C1P1-virus strain and VE6T cells (Geometric mean titer (GMT) 133) than with the VE6-adapted FIN-1 in either cell line (GMT 53 with VE6-cells and 66 with VE6T cells) (p < 0.001). There were no differences in NAb titers against VE6-adapted virus strain in VE6T compared to VE6 cells (p = 0.685) (Table S3). The overall higher titers of the tested samples on VE6T cells with the C1P1 strain and the differences between the cell culture systems suggest that comparisons between variant strains should preferably be done using cell lines with relevant molecules affecting entry and representative low-passage clinical isolates of the strains. Consequently, further comparisons to variant strains were performed with the C1P1 and VE6T cells.
3.2. Overall NAb Titers against C1P1 Compared to Variant Strains
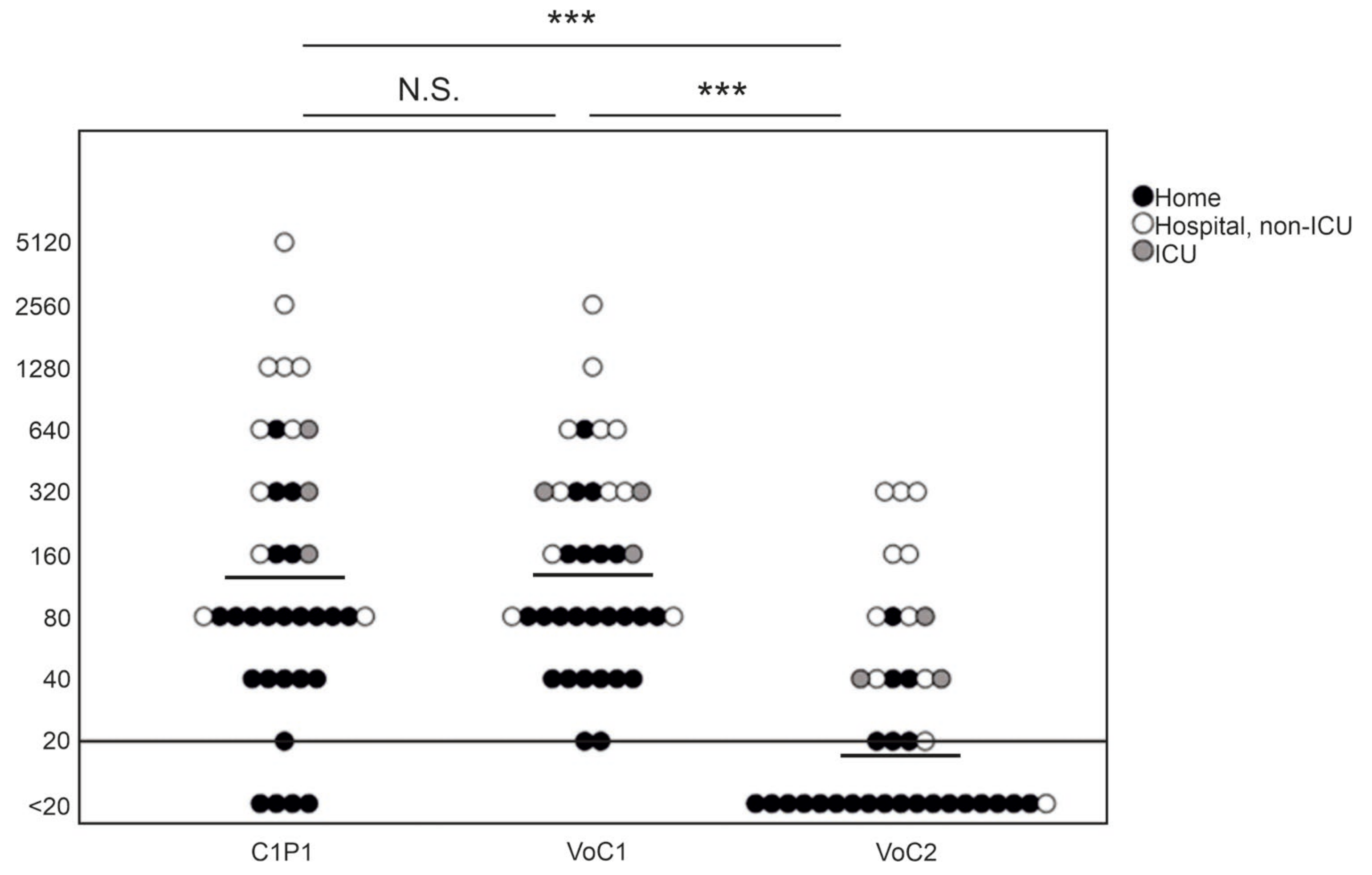
NAb titers against C1P1 were compared to VoC1 and VoC2 using VE6T cells (Figure 1, Table S3). The GMT was 141 with the VoC1 strain, and 17 with the VoC2 strain, as compared to 133 with the C1P1. The titers were significantly lower, with an approximately 8-fold lower GMT against VoC2 as compared to both VoC1 and C1P1 (p < 0.001). Titers of four samples remained below the first tested dilution (<20) with the C1P1 strain, 19 with the VoC2 strain, and none with the VoC1 strain. There was no statistically significant difference between VoC1 and C1P1 (p = 1.000).
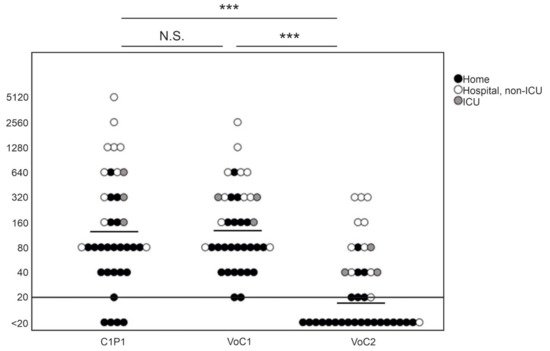
Figure 1.
Comparison of neutralizing antibody titers with geometric mean lines against different SARS-CoV-2 strains. Comparison of C1P1, VoC1, and VoC2 titers with individual data points indicated in the picture. Titers are expressed in logarithmic scale (Log2) and LOD has been marked with a horizontal line. Statistical significances are indicated with *** (p < 0.001) and N.S. (p > 0.05).
3.3. Comparison of IgG and NAb Titers
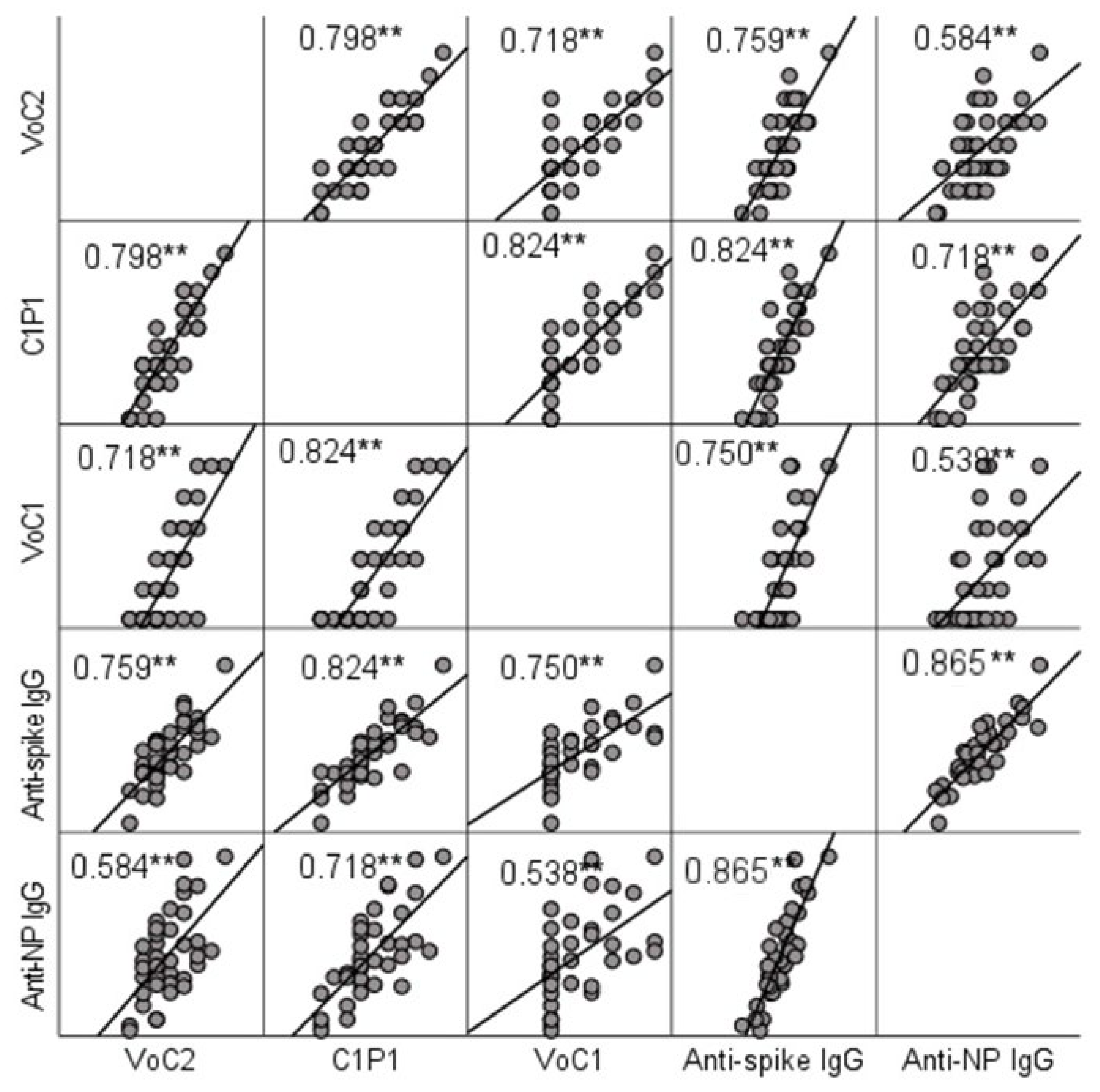
NAb titers were compared to IgG titers determined by ELISA. A significant positive correlation (p < 0.001) with Spearman’s rho-values ranging between 0.584 (anti-NP and VoC2) and 0.824 (anti-spike and C1P1) was found between MNT and ELISA result in all cases (Figure 2, Table S3). Anti-spike-IgG and anti-nucleoprotein-IgG titers did not show a significant difference (p = 0.960).
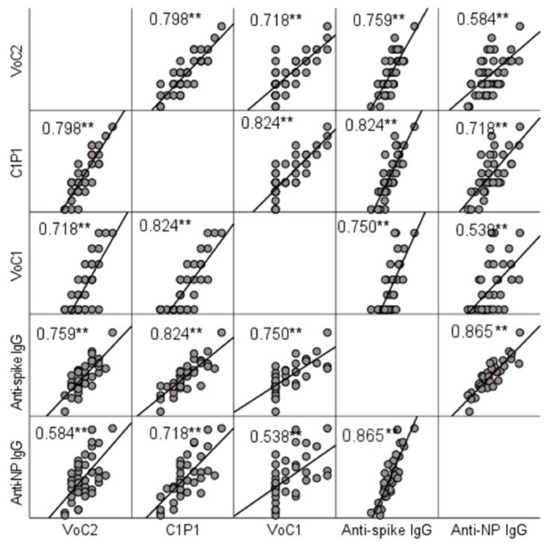
Figure 2.
Correlation between NAb titers and IgG titers determined by ELISA testing. Scatter matrix comparing NAb titers with 3 virus strains and IgG titers with spike protein and nucleoprotein. Spearman’s rho-values between NAb titers with each virus strain and anti-spike and anti-NP IgGs are included in the picture, and significant values at level 0.01 (2-tailed) are marked with **.
3.4. NAb Titers in Relation to Time from the Disease and Disease Severity
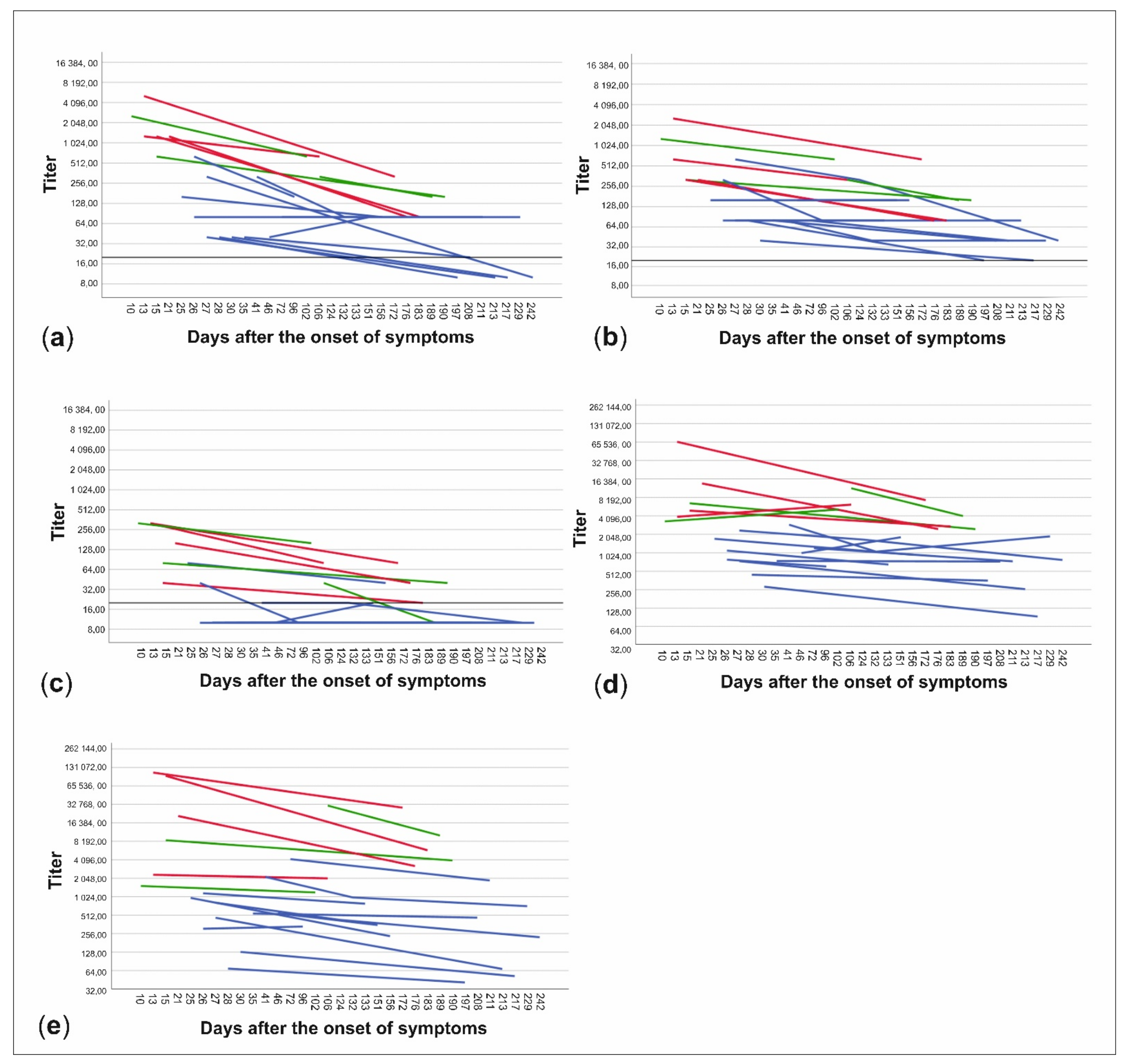
The patient data were further divided into subgroups based on whether the patients had been treated at home or had needed hospital care and on the other hand, whether the time from onset of symptoms to date of serum sampling was less than or more than 150 days. NAb titers to all three virus strains and anti-spike- and anti-NP-IgG ELISA titers were higher in patients treated in hospital than in those treated at home (p < 0.001) (Figure 3, Table 1). Titers were also higher in the sera drawn less than 150 days from the onset of illness in all cases, but the difference was statistically significant only with C1P1 (p = 0.007) and VoC1 (p = 0.012) (Table 1, Figure 3, and Table S3). Using VoC2, 5/7 samples taken between days 150–200 were NAb-positive but none of the 6 samples taken after 200 days had detectable levels of NAbs, whereas with C1P1 and VoC1, a large proportion of the patients still had detectable levels of NAbs 200 days after the onset of disease.
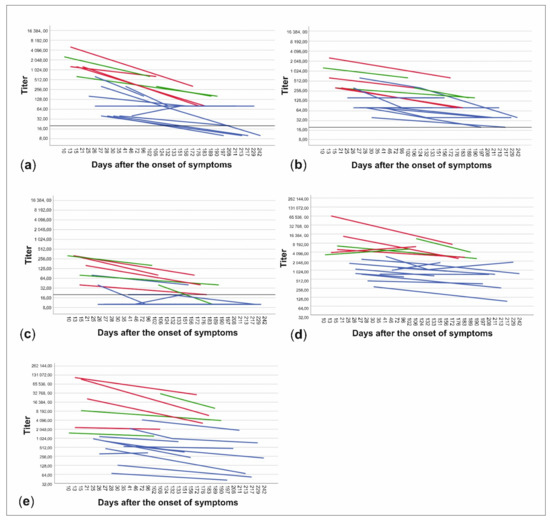
Figure 3.
Changes in neutralizing antibody titers and anti-spike- and anti-NP-IgG titers between samples of each patient. Titers are expressed as neutralizing antibody titers for C1P1 (a), VoC1 (b), and VoC2 (c) as well as end-point titers for anti-spike- (d) and anti-NP-IgG (e) antibodies. Patients treated at home are shown in blue and patients treated in the hospital are shown in red (non-ICU) and green (ICU). Titers are depicted in logarithmic scale (Log2), LOD has been marked with a horizontal line, and titers below LOD in A-C have been set to ten.

Table 1.
Geometric mean titers and p-values when the data is divided into subgroups based on the time after the onset of symptoms and disease severity.
Slopes of fit lines between paired samples of individual patients were used to compare the rates of NAb titer decrease between virus strains. The average slope value was slightly more negative (i.e., the descending slope was steeper) with C1P1 (−1.1 × 10−2, SD 1.1 × 10−2) than VoC1 (−9.41 × 10−3, SD 9.02 × 10−3), or VoC2 (−9.90 × 10−3 SD 4.84 × 10−3), but differences were not statistically significant with Friedman’s analysis of variance by ranks test (p = 0.186) or in pairwise comparisons with the Wilcoxon signed rank test (Table S3), suggesting that the waning of the NAb titers occurred at similar pace towards the different variants.
4. Discussion
The emergence of new VoC strains of SARS-CoV-2 requires active research to understand their phenotypic and antigenic properties of relevance for the pandemic and its control. This study contributes to the gathering of information on the neutralizing antibody kinetics of previously infected individuals towards clinical isolates representing different strains of SARS-CoV-2. The patients from which the paired samples in this study were collected with high likelihood experienced D614G infection in spring 2020 [25]. These strains are represented by the C1P1 strain, whereas now (April 2021) in Finland the B.1.1.7 strain represents ca. 70% and B.1.351 10–20% of circulating strains. This is why these strains isolated from Finnish patients were selected for MNT analyses.
The obtained results are in line with previous studies reporting lowered NAb levels against the VoC2 (B.1.351) variant when compared to VoC1 (B.1.1.7) or the older dominant strains, including studies in vaccinees [5,8,11,13,26,27]. It should be noted that a large proportion of VoC2 titers were under the detection limit of 20 and were set to value of 10 for calculations, which means that statistical values of VoC2 are not precise. However, the significance of the difference between strains can be judged to be reliable and the relatively high value of 10 was used to avoid reporting false significance. The weak NAb immunity of the first wave recoverees towards VoC2 suggest that the latter lineage poses a public health threat for early re-emergence of SARS-CoV2 in previously infected populations. The results provide additional evidence justifying measures to control the spread of VoC2 and other lineages with immune escape properties.
The neutralizing response towards the contemporary and VoC1 strains, although waning, was shown to last for months. No differences were detected in the relative rate in which the titer towards each virus strain decreased within this dataset. One should note, however, that due to the large number of samples excluded from this comparison because of the titers below the detection limit, the sample size in the analysis was small. Furthermore, the decrease in NAb response might not be linear in the 2-log scale as was assumed here. In any case, none of the samples taken over 200 days after disease onset showed detectable levels of NAbs against VoC2, which can be explained by the significantly lower initial levels. This indicates that protection provided by NAb response from an infection with strains circulating in 2020 does not last as long against VoC2 as it does against C1P1 and VoC1.
ELISA and NAb titers correlated well and this suggests that ELISA could serve as a rapid and convenient screening method for identifying high NAb level patients. As in other studies [28,29,30,31], significantly higher antibody responses in both ELISA and NAb tests were measured in patients who needed hospital care as compared to those requiring only outpatient care. The individual NAb responses also showed the presence of occasional “pan-reactive” recoverees with high NAb titers to all variants (Table S1), which could serve as potential donors for e.g., memory B cells for cloning antibodies for therapeutic purposes.
Although previous studies have shown that in general the different types of SARS-CoV-2 neutralization assays have good concordance [32,33], comparisons of different assays are needed for evaluating the properties of different circulating virus strains that represent naturally occurring sets of mutations. The current understanding of SARS-CoV-2 NAb kinetics is based on results obtained using various techniques, cell lines, and virus strains [6]. Our observation that the TMPRSS2-expressing cells made a VE6 cell-based microneutralization test more sensitive translates to an overall better longevity of the antibody response, and could imply that the results obtained using different assay protocols providing heterologous entry molecules for the virus may vary, calling for further standardization of the NAb assays [5,34].
In conclusion, our results support that the strains largely circulating in 2020 provide sustained antibody-mediated protection towards the contemporary strains as well as the B.1.1.7 variant of concern rapidly spreading in e.g., Europe and the United States, but poorly against the B.1.351 variant, explaining its potential for surge in previously infected populations and highlighting the need for control measures to prevent this variant from further spread [35,36,37].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13060996/s1, Table S1: Sample information and results. Table S2: Mutations of virus strains used in the study compared to strain Wuhan Hu-1. Table S3: Statistical tests and parameters.
Author Contributions
Conceptualization, O.V., J.V., E.M.K., K.A., R.U., and T.S. (Tarja Sironen); methodology, J.V., V.N., E.M.K., K.A., R.U., J.H., T.S. (Teemu Smura), and S.K.; investigation, J.V., V.N., E.M.K., K.A., R.U., S.H.P., S.M., A.P., M.R., T.S. (Teemu Smura), J.H., and S.K.; resources, O.V., A.K., and T.S. (Tarja Sironen); data curation, S.H.P., S.M., A.P., M.R., J.V., R.U., K.A., E.M.K., T.S. (Teemu Smura), S.K., and V.N.; writing—original draft preparation, J.V., E.H., O.V., E.M.K., R.U., and K.A.; writing—review and editing, O.V., E.M.K., J.V., R.U., K.A., E.H., J.H., T.S. (Teemu Smura), and S.K.; visualization, J.V. and R.U.; supervision, K.H., O.V., A.K., and T.S. (Tarja Sironen); project administration, O.V. and E.H.; funding acquisition, K.H. and O.V. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Jane and Aatos Erkko Foundation, EU Horizon (VEO, 874735), Academy of Finland and Helsinki University Hospital Funds (TYH2018322).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and was approved by the ethical committee of the Department of Medicine at the Helsinki University Hospital (HUS/853/2020, approved 25 March 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in the article and in its online Supplementary Materials.
Acknowledgments
We thank Esa Pohjolainen, Taru Miller, Simo Miettinen, Dina Mosselhy, and Lea Hedman for their expert technical assistance, and staff at HUSLAB Virology and Immunology for providing samples for virus isolation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Addetia, A.; Crawford, K.H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.L.; Jerome, K.R.; Bloom, J.D.; Greninger, A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, e02107–e02120. [Google Scholar] [CrossRef] [PubMed]
- Alsoussi, W.B.; Turner, J.S.; Case, J.B.; Zhao, H.; Schmitz, A.J.; Zhou, J.Q.; Chen, R.E.; Lei, T.; Rizk, A.A.; McIntire, K.M.; et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020, 205, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371, eabf4063. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393. [Google Scholar] [CrossRef]
- Li, Q.; Nie, J.; Wu, J.; Zhang, L.; Ding, R.; Wang, H.; Zhang, Y.; Li, T.; Liu, S.; Zhang, M.; et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 2021, 184, 2362–2371. [Google Scholar] [CrossRef]
- Rees-Spear, C.; Muir, L.; Griffith, S.A.; Heaney, J.; Aldon, Y.; Snitselaar, J.L.; Thomas, P.; Graham, C.; Seow, J.; Lee, N.; et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021, 34, 108890. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef]
- Tada, T.; Dcosta, B.M.; Samanovic-Golden, M.; Herati, R.S.; Cornelius, A.; Mulligan, M.J.; Landau, N.R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv 2021, 7, 2021.02.05.430003. [Google Scholar] [CrossRef]
- Pısıl, Y.; Shida, H.; Miura, T. A Neutralization Assay Based on Pseudo-Typed Lentivirus with SARS CoV-2 Spike Protein in ACE2-Expressing CRFK Cells. Pathogens 2021, 10, 153. [Google Scholar] [CrossRef]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Zettl, F.; Meister, T.L.; Vollmer, T.; Fischer, B.; Steinmann, J.; Krawczyk, A.; V’kovski, P.; Todt, D.; Steinmann, E.; Pfaender, S.; et al. Rapid Quantification of SARS-CoV-2-Neutralizing Antibodies Using Propagation-Defective Vesicular Stomatitis Virus Pseudotypes. Vaccines 2020, 8, 386. [Google Scholar] [CrossRef]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856. [Google Scholar] [CrossRef]
- Rusanen, J.; Kareinen, L.; Szirovicza, L.; Uğurlu, H.; Levanov, L.; Jääskeläinen, A.; Ahava, M.; Kurkela, S.; Saksela, K.; Hedman, K.; et al. A Generic, Scalable, and Rapid Time-Resolved Förster Resonance Energy Transfer-Based Assay for Antigen Detection-SARS-CoV-2 as a Proof of Concept. mBio 2021, 12, e00902-21. [Google Scholar] [CrossRef]
- Haveri, A.; Smura, T.; Kuivanen, S.; Österlund, P.; Hepojoki, J.; Ikonen, N.; Pitkäpaasi, M.; Blomqvist, S.; Rönkkö, E.; Kantele, A.; et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance 2020, 25, 2000266. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Stadlbauer, D.; Amanat, F.; Chromikova, V.; Jiang, K.; Strohmeier, S.; Arunkumar, G.A.; Tan, J.; Bhavsar, D.; Capuano, C.; Kirkpatrick, E.; et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57, e100. [Google Scholar] [CrossRef]
- Rusanen, J.; Kareinen, L.; Levanov, L.; Mero, S.; Pakkanen, S.H.; Kantele, A.; Amanat, F.; Krammer, F.; Hedman, K.; Vapalahti, O.; et al. A 10-Minute "Mix and Read" Antibody Assay for SARS-CoV-2. Viruses 2021, 13, 143. [Google Scholar] [CrossRef] [PubMed]
- Alm, E.; Broberg, E.K.; Connor, T.; Hodcroft, E.B.; Komissarov, A.B.; Maurer-Stroh, S.; Melidou, A.; Neher, R.A.; O’Toole, Á.; Pereyaslov, D. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance 2020, 25, 2001410. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Sakharkar, M.; Rappazzo, C.G.; Wieland-Alter, W.F.; Hsieh, C.L.; Wrapp, D.; Esterman, E.S.; Kaku, C.I.; Wec, A.Z.; Geoghegan, J.C.; McLellan, J.S.; et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabg6916. [Google Scholar] [CrossRef]
- Batra, M.; Tian, R.; Zhang, C.; Clarence, E.; Sacher, C.S.; Miranda, J.N.; De La Fuente, J.R.O.; Mathew, M.; Green, D.; Patel, S.; et al. Role of IgG against N-protein of SARS-CoV2 in COVID19 clinical outcomes. Sci. Rep. 2021, 11, 3455. [Google Scholar] [CrossRef]
- Graham, N.R.; Whitaker, A.N.; Strother, C.A.; Miles, A.K.; Grier, D.; McElvany, B.D.; Bruce, E.A.; Poynter, M.E.; Pierce, K.K.; Kirkpatrick, B.D.; et al. Kinetics and isotype assessment of antibodies targeting the spike protein receptor-binding domain of severe acute respiratory syndrome-coronavirus-2 in COVID-19 patients as a function of age, biological sex and disease severity. Clin. Transl. Immunol. 2020, 9, e1189. [Google Scholar] [CrossRef]
- Long, Q.; Tang, X.; Shi, Q.; Li, Q.; Deng, H.; Yuan, J.; Hu, J.; Xu, W.; Zhang, Y.; Lv, F.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Wang, F.; Li, X.; Liang, X.; Zhou, Y.; Zhang, D.; Liu, J.; Zeng, H.; Wang, J.; Shi, Y. Correlation of the ratio of IgM/IgG concentration to days after symptom onset (IgM/T or IgG/T) with disease severity and outcome in non-critical COVID-19 patients. Am. J. Transl. Res. 2021, 13, 1197–1208. [Google Scholar] [PubMed]
- Riepler, L.; Rössler, A.; Falch, A.; Volland, A.; Borena, W.; von Laer, D.; Kimpel, J. Comparison of Four SARS-CoV-2 Neutralization Assays. Vaccines 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef]
- Harvala, H.; Mehew, J.; Robb, M.L.; Ijaz, S.; Dicks, S.; Patel, M.; Watkins, N.; Simmonds, P.; Brooks, T.; Johnson, R.; et al. Convalescent plasma treatment for SARS-CoV-2 infection: Analysis of the first 436 donors in England, 22 April to 12 May 2020. Eurosurveillance 2020, 25, 2001260. [Google Scholar] [CrossRef]
- Betton, M.; Livrozet, M.; Planas, D.; Fayol, A.; Monel, B.; Védie, B.; Bruel, T.; Tartour, E.; Robillard, N.; Manuguerra, J.C.; et al. Sera neutralizing activities against SARS-CoV-2 and multiple variants six month after hospitalization for COVID-19. Clin. Infect. Dis. 2021, ciab308. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Hodcroft, E.B.; Fauver, J.R.; Phelan, A.L.; Cevik, M. Public health actions to control new SARS-CoV-2 variants. Cell 2021, 184, 1127–1132. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).