Abstract
P3N-PIPO (P3 N-terminal fused with Pretty Interesting Potyviridae ORF), the movement protein of potyviruses, is expressed as a translational fusion with the N-terminus of P3 in potyviruses. As reported in previous studies, P3N-PIPO is expressed via transcriptional slippage at a conserved G2A6 slippery site in the genus Potyvirus. However, it is still unknown whether a similar expression mechanism of P3N-PIPO is used in the other genera of the family Potyviridae. Moreover, due to the extremely low expression level of P3N-PIPO in natural virus-infected plants, the peptides spanning the slippery site which provide direct evidence of the slippage at the protein level, have not been identified yet. In this study, a potato virus X (PVX)-based expression vector was utilized to investigate the expression mechanism of P3N-PIPO. A high expression level of the P3N-PIPO(WT) of turnip mosaic virus (TuMV, genus Potyvirus) was observed based on the PVX expression vector. For the first time, we successfully identified the peptides of P3N-PIPO spanning the slippery site by mass spectrometry. Likewise, the P3N-PIPO(WT) of wheat yellow mosaic virus (WYMV, genus Bymovirus) was also successfully expressed using the PVX expression vector. Integrated proteome and transcriptome analyses revealed that WYMV P3N-PIPO was expressed at the conserved G2A6 site through transcriptional slippage. Moreover, as revealed by mutagenesis analysis, Hexa-adenosine of the G2A6 site was important for the frameshift expression of P3N-PIPO in WYMV. According to our results, the PVX-based expression vector might be used as an excellent tool to study the expression mechanism of P3N-PIPO in Potyviridae. To the best of our knowledge, this is the first experimental evidence dissecting the expression mechanism of a bymovirus P3N-PIPO in the experimental host Nicotiana benthamiana.
1. Introduction
Most RNA viruses have a relatively small genome size, which seems to be insufficient for RNA viruses to cope with long-term selective pressure. RNA virus has explored various strategies to maximize its coding capacities, such as splicing, transcriptional slippage, ribosomal frameshifting, leaky scanning, and stop-codon read-through [1,2].
Amongst them, transcriptional slippage often takes place on repetitive nucleotides such as poly(A) or poly(T) tracts, resulting in the synthesis of heterogeneous mRNAs that always insert one or more extra nucleotides but seldom delete one or two bases [3]. If this occurs in a coding sequence, then the heterogeneous mRNA will translate into more than one protein product. It is well-recognized that transcriptional slippage is utilized in bacteria such as Escherichia. Coli [3,4]. For example, the expressions of E. coli operons, pyrBI and codBA, were regulated by transcriptional slippage under special conditions [5,6]. As for animal viruses, transcriptional slippage has also been observed in the Ebola virus [7,8,9] (a negative-strand RNA virus) and the Hepatitis C virus (HCV) [10] (a positive-strand RNA virus). With regard to plant viruses, replication slippage(transcriptional slippage)is considered a common evolution process in the genus Potyvirus [11], but direct evidence is still missing due to the extremely low expression level of the transcriptional slippage during infection [12].
A short open reading frame (ORF), known as PIPO (Pretty Interesting Potyviridae ORF), is discovered within the genome sequences of Potyviridae by bioinformatics analysis [13]. PIPO was proven to be expressed as a part of a fusion protein with the P3 N-terminal region, named P3N-PIPO [13,14]. Thereafter, numerous studies have been conducted, and the results show that P3N-PIPO is the movement protein of potyviruses [14,15,16,17,18]. High-throughput sequencing has shown that transcriptional slippage at the G2A6 site accounts for the expression mechanism of P3N-PIPO in potyviruses [2,19]. It is worth noting that previous studies on P3N-PIPO mainly focused on the genus Potyvirus, but there are no experimental data supporting whether this mechanism is widely used by the other members in the family Potyviridae. Moreover, previous studies suggested that the slippage efficiency varied between 0.8 and 2.1% in potyviruses [2,19]. Owing to the extremely low expression level of P3N-PIPO in natural virus-infected plants, a reliable experimental system is needed to isolate sufficient frameshift-derived protein for analysis. The potato virus X (PVX)-based expression vectors are commonly used exogenous protein expression systems due to their systemic expression and high expression levels in plants [20,21].
In this study, the PVX-based vector system was utilized to investigate the expression mechanism of P3N-PIPO. The frameshift expression of P3N-PIPO(WT)-GFP was successfully identified using the PVX-based vector in plants. In addition, the frameshift of P3N-PIPO(WT) in another plant virus, wheat yellow mosaic virus (WYMV) of the genus Bymovirus, was also successfully identified and characterized.
Our research not only sheds more light on the expression mechanism of P3N-PIPO in the family Potyviridae, but also provides an excellent tool to study the slippage efficiency of P3N-PIPO.
2. Materials and Methods
2.1. Construction of PVX-Derived Vectors
The PVX vector (pgR106) used in this study was obtained from David Baulcombe’s laboratory. A gfp fragment without the start codon was amplified from a GFP binary vector using specific oligonucleotide primers (Table S1) that incorporated AscI/SnaBI and SalI restrictions sites at the 5′- and 3′-terminal, respectively. Then, this gfp fragment was cloned into the pgR106 vector in line with the standard molecular clone protocols and was named PVX-GFP (AscI/SnaBI, SalI).
Additionally, P3N-PIPO(WT) was amplified from TuMV and WYMV infectious clones, respectively, by the use of specific oligonucleotide primers (Table S1) that incorporated ClaI and SnaBI restrictions sites at the 5′- and 3′-terminus, respectively. Afterwards, this fragment was cloned into the pJET1.2/blunt vector (Thermo Fisher Scientific, Waltham, MA, USA) following the standard molecular clone protocols and was named the pJET-P3N-PIPO (ClaI/SnaBI) vector.
The in-frame control construct WYMV P3N-PIPO(FS-1)-GFP was generated by inserting adenosine into the G2A6 site of WYMV P3N-PIPO(WT) with the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies Inc., Palo Alto, CA, USA), according to the manufacturer’s instructions (Table S1). Other mutants like PVX-WYMV-P3N-PIPO-M1 and PVX-WYMV-P3N-PIPO-M2 were constructed by the same method (Table S1).
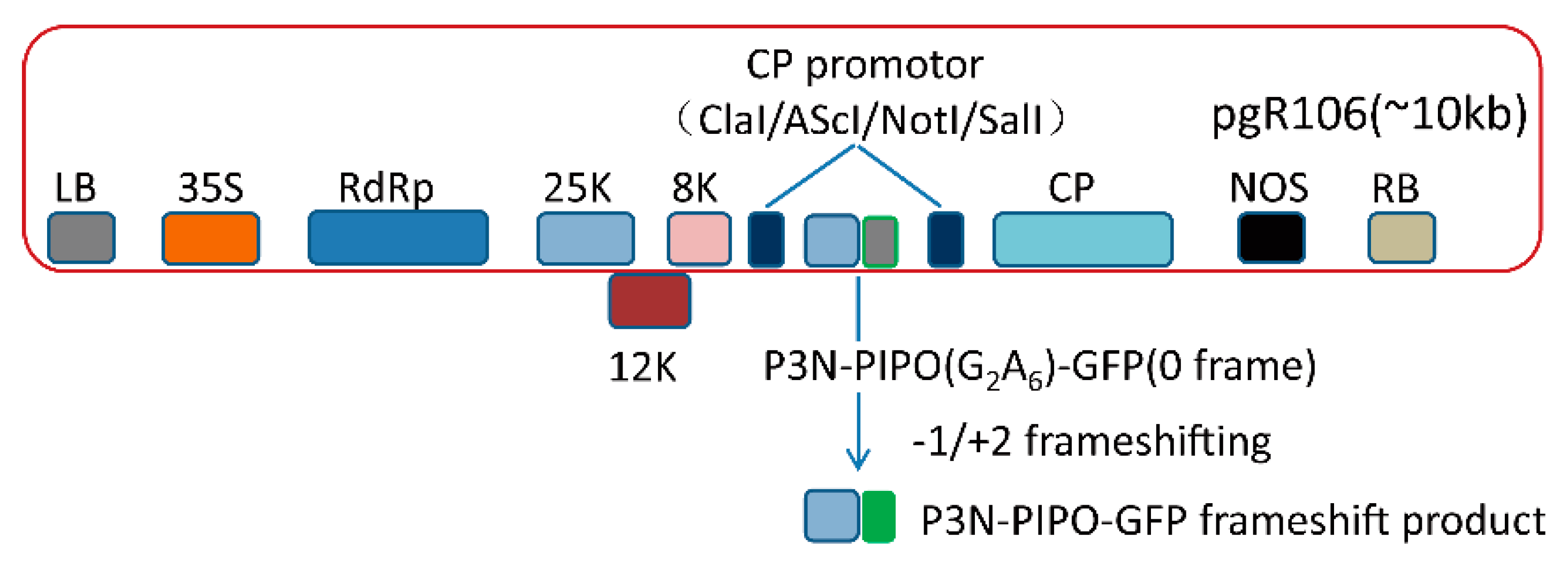
First of all, PVX-GFP (AscI/SnaBI, SalI) and pJET-P3N-PIPO (ClaI/SnaBI) were digested with ClaI and SnaBI, respectively. Then, PVX-GFP (ClaI/SnaBI) and P3N-PIPO (ClaI/SnaBI) were purified by gel extraction (Thermo Fisher Scientific, Waltham, MA, USA) and ligated by T4 DNA ligase (Figure 1).
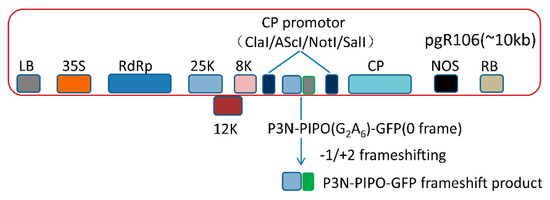
Figure 1.
The schematic diagram of P3N-PIPO frameshift expression vector.
2.2. Growth of Nicotiana Benthamiana Plants, Agrobacterium Infection of Plants, and Confocal Imaging Analysis
N. benthamiana were grown in a glasshouse under the following conditions: temperature (day: night) 25 °C: 22 °C ± 2 °C, and 16 h of light.
All the PVX chimeric virus vectors constructed in this study were transformed into the Agrobacterium tumefaciens strain GV3101 (carrying the helper plasmid pSoup), as described previously [22]. Moreover, agroinfiltration of N. benthamiana with PVX chimeric virus vectors was also implemented, as described previously [23].
Plant leaves expressing recombinant proteins were imaged using a Leica TCS SP2 confocal microscope. Green fluorescent protein (GFP) was excited at 488 nm, and the emitted light was captured at 507 nm. Images were captured digitally and processed using the Leica LCS software.
2.3. Western Blotting (WB) Analysis
To analyze the frameshift expression of P3N-PIPO, total proteins were extracted from N. benthamiana leaves infected with PVX chimeric viruses using a lysis buffer (100 mM Tris-HCl (pH = 6.8), 20% SDS, and 2% β-mercaptoethanol) and were separated by 12% SDS-PAGE. Proteins were electroblotted onto a preactivated PVDF membrane. Then, the membrane was blocked with block buffer (50 mL 1 × TBS buffer containing 5% skimmed milk powder) and probed with the GFP monoclonal antibody (Sigma-Aldrich Corp., St. Louis, MO, USA) at room temperature for 2 h. Following three washes of 5 min each in 1 × TBS containing 0.1% Tween 20, proteins were incubated with alkaline phosphatase (AP)-conjugated polyclonal goat anti-mouse IgG (Sigma-Aldrich Corp., St. Louis, MO, USA) and visualized using the NBT/BCIP® solution (Sigma-Aldrich Corp., St. Louis, MO, USA).
2.4. Protein Purification
All GFP-fused proteins were purified by the GFP-TRAP-A purification kit (Chromo Tek, Rosemont, IL, USA) following the manufacturer’s instructions. Thereafter, the GFP-TRAP bound fraction was resolved by SDS-PAGE and stained with the EZBlue™ gel staining reagent (Sigma-Aldrich Corp., St. Louis, MO, USA).
2.5. Mass Spectrometric Analysis
Proteins of interest were excised from the stained SDS-PAGE gels, followed by in-gel digestion with trypsin, and analysis by nano-LC/MS/MS according to standard protocols. Samples were prepared as previously described by Granvogl et al. [24]. Mass spectrometry was carried out at the fingerprints proteomics facility of the University of Dundee and the Biomedical Sciences Research Complex mass spectrometry and proteomics facility of the University of St Andrews.
2.6. High Throughput Sequencing
The infiltration patches were harvested eight days after the infiltration of N. benthamiana with viruses. Total RNA was extracted from the collected samples with TRIzol reagent (Invitrogen, Waltham, MA, USA) in line with the manufacturer’s instructions. Sequencing libraries were prepared using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s recommendations. Meanwhile, index codes were added to attribute sequences to each sample. The library quality was assessed using the Agilent Bioanalyzer 2100 system. Deep sequencing was performed on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA, USA) by Tianjin Novogene Bioinformatic Technology Co., Ltd. (Tianjin, China).
Raw reads of the viral vector from Hiseq 4000 sequencing were quality trimmed. The remaining reads were mapped to P3N-PIPO sequences of TuMV and WYMV using bowtie2 (version 2.4.1) with default parameters. Later, the mapped reads to identify insertion/deletion (indel) were performed by custom Perl scripts and Linux shell bash scripts.
3. Results
3.1. Frameshift Expression of P3N-PIPO in N. benthamiana by PVX-Based Vector
To test whether the PVX-based vectors were suitable for P3N-PIPO frameshift expression or not, we inserted the WYMV P3N-PIPO(WT) and TuMV P3N-PIPO(WT) into the PVX-based vector pgR106, which then named PVX-GFP (AscI/SnaBI; SalI) (Figure 1). It was expected that the frameshift expression of the constructs would result in the trans-frame fused expression of P3N-PIPO-GFP, in which GFP fluorescence was detected under a fluorescence microscope.
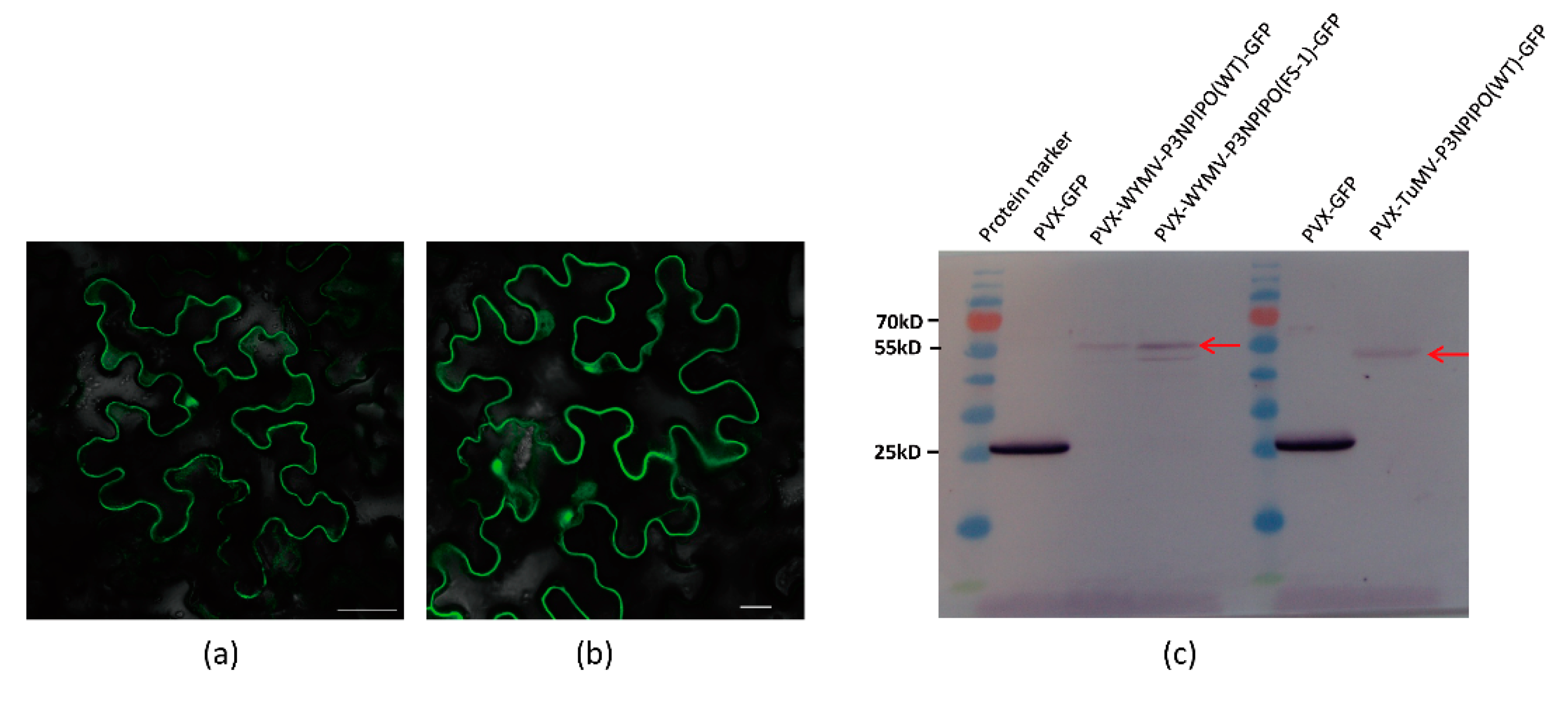
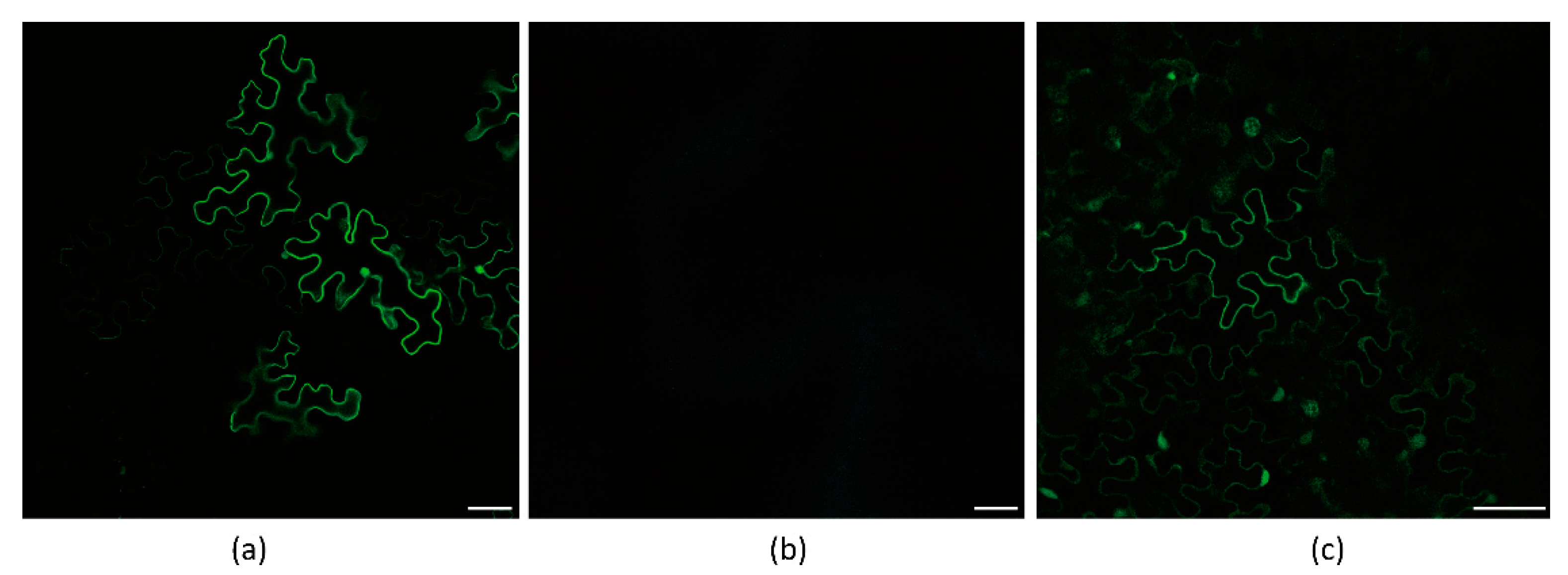
As a result, these constructs were expressed in N. benthamiana, and the fluorescence of P3N-PIPO-GFP was successfully detected by Laser Scanning Confocal Microscopy (LSCM) (Figure 2a,b).
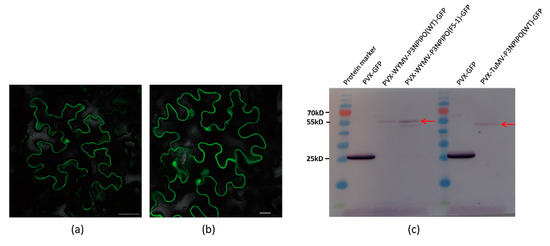
Figure 2.
Frameshift expression of P3N-PIPO(WT) in N. benthamiana leaf cells. (a) PVX-TuMV-P3N-PIPO(WT)-GFP expressed in the epidermal cells treated eight days post-agroinfiltration. (b) PVX-WYMV-P3N-PIPO(WT)-GFP expressed in the epidermal cells treated eight days post-agroinfiltration. Bars, 50 μM. (c) Immunodetection of the frameshift expression of PVX-TuMV-P3N-PIPO(WT)-GFP and PVX-WYMV-P3N-PIPO(WT)-GFP in N. benthamiana.
To further confirm the expression of P3N-PIPO-GFP, we utilized a GFP antibody to detect the frameshift expression of P3N-PIPO. Additionally, an in-frame control, where the predicted shift site GGA_AAA_AAT_C was mutated to GGA_AAA_AAA_T to force the expression of WYMV P3N-PIPO(FS-1)-GFP, was prepared to indicate the approximate size (56 kD) at which the frameshift protein should theoretically migrate in the gels. Using PVX-GFP as a positive control, the frameshift expression of WYMV P3N-PIPO(WT)-GFP and TuMV P3N-PIPO(WT)-GFP was successfully detected (Figure 2c).
3.2. Mass Spectrometry Revealed That Frameshifting Used in P3N-PIPO(WT)-GFP Expression Took Place at the G2A6 Site
To determine the precise sites and direction of the frameshift expression of P3N-PIPO, total proteins were extracted from N. benthamiana infected with PVX-TuMV-P3N-PIPO(WT)-GFP and PVX-WYMV-P3N-PIPO(WT)-GFP. Later, the frameshift proteins were affinity-purified by GFP-TRAP beads and separated through SDS-PAGE. Afterwards, gel slices containing frameshift proteins were excised and digested with trypsin and were analyzed by mass spectrometry.
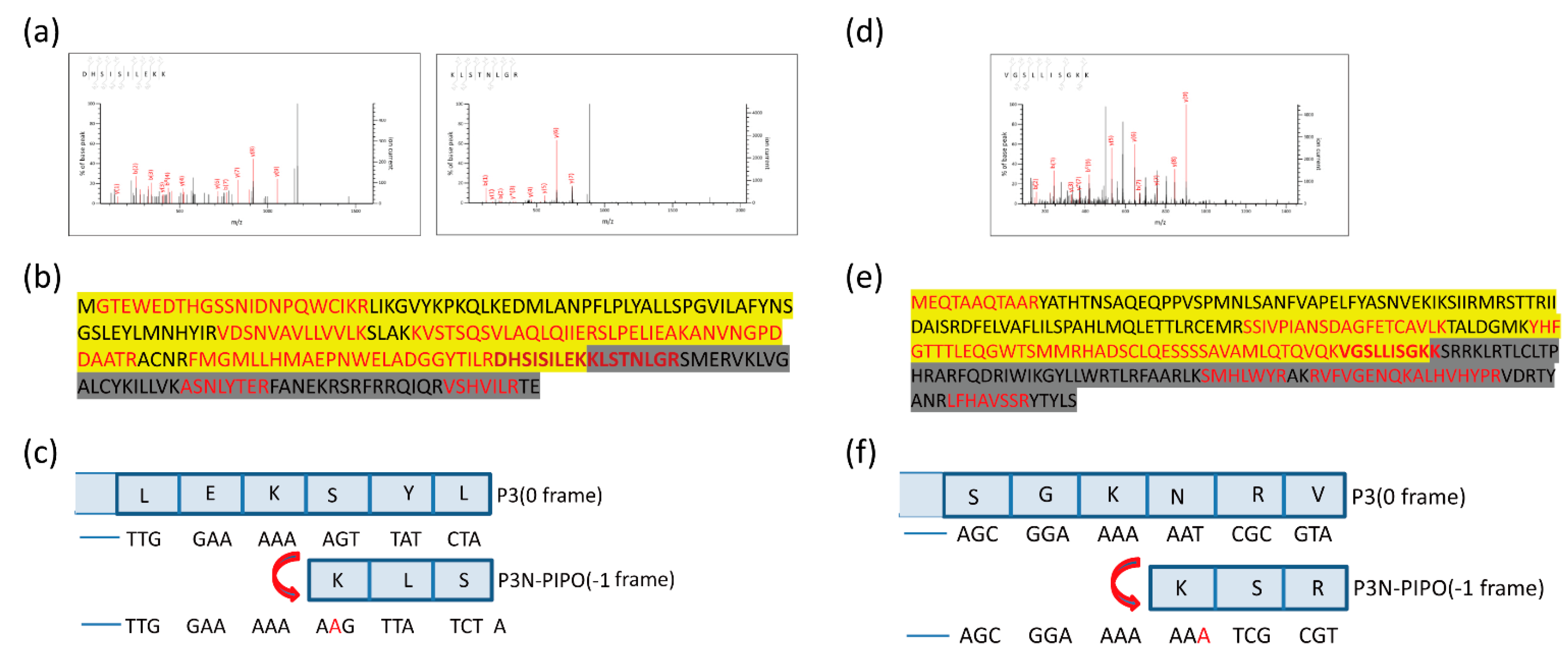
Altogether, 10 unique peptides of TuMV P3N-PIPO and nine unique peptides of WYMV P3N-PIPO were identified, including peptides located in both the upstream and downstream predicted shift sites (Figure 3 and File S1). As the predicted shift site contains the trypsin digestion sites (arginine and lysine), it is difficult to obtain the tryptic peptide containing the predicted shift site. If there is an acidic amino like aspartic acid or glutamic acid in the direct neighborhood of the digestion site, the rate of hydrolysis is diminished [25]. Fortunately, glutamic acid was found near the digestion site at the predicted shift site of TuMV P3N-PIPO-GFP, suggesting the successful identification of two peptides spanning the shift site (Figure 3a). The peptides DHSISILEKK and KLSTNLGR showed that the frameshifting used in TuMV P3N-PIPO-GFP expression occurred at the G2A6 site in the –1 directions (Figure 3c). Another peptide of WYMV P3N-PIPO-GFP, VGSLLISGKK, contained the shift site, which showed that the frameshifting used in WYMV P3N-PIPO-GFP expression took place at the G2A6 site as well (Figure 3d,f).
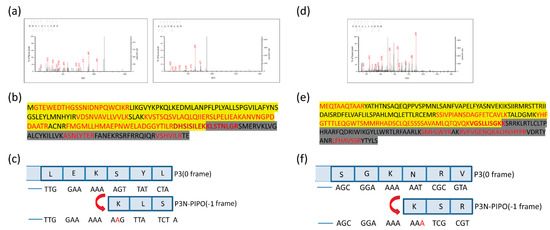
Figure 3.
Mass spectrometry of the PVX-TuMV-P3N-PIPO(WT)-GFP and PVX-WYMV-P3N-PIPO(WT)-GFP frameshift expression proteins. (a) MS/MS fragmentation spectra of the shift site peptides DHSISILEKK and KLSTNLGR in TuMV-P3N-PIPO(WT). (b) The complete amino acid sequence of TuMV P3NPIPO. The peptides before the G2A6 site are highlighted in yellow. Amino acids encoded by the –1 frame are highlighted in dark. (c) Nucleotide sequence around the G2A6 shift site, and conceptual amino acids translated in the 0 and –1 reading frames in TuMV-P3N-PIPO. (d) MS/MS fragmentation spectrum of the shift site peptide VGSLLISGKK in WYMV-P3N-PIPO(WT). (e) The complete amino acid sequence of WYMV P3NPIPO. The peptides before the G2A6 site are highlighted in yellow. Amino acids encoded by the −1 frame are highlighted in dark. (f) Nucleotide sequence around the G2A6 shift site, and conceptual amino acids translated in the 0 and −1 reading frames in WYMV-P3N-PIPO.
3.3. Deep Sequencing Confirmed the Adenosine Insertion at the G2A6 Site of P3N-PIPO
To further investigate the expression mechanism of P3N-PIPO at the transcriptional level, a transcriptomics analysis of PVX-P3N-PIPO-infected plants was performed by high-throughput sequencing.
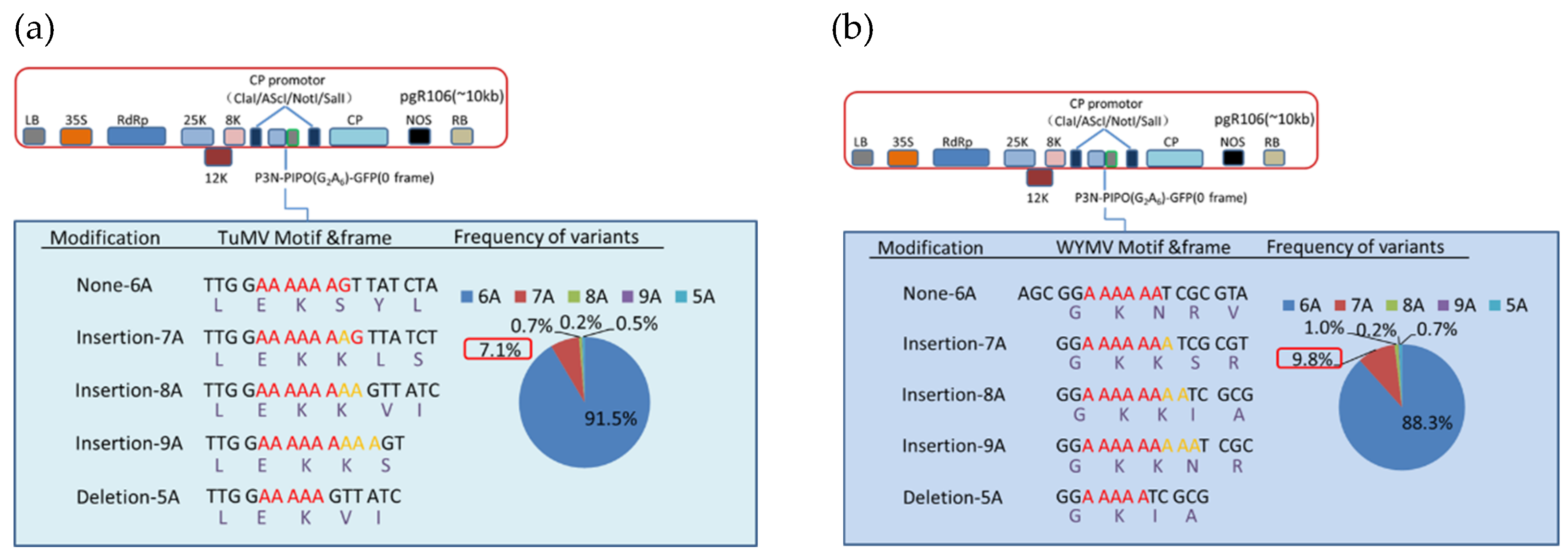
According to our results, various deletions and insertions of adenosine could be detected at the G2A6 site of P3N-PIPO of TuMV and WYMV. Besides, single adenosine insertion was the most abundant transcript editing form, and there was 7.1% and 9.8% of the transcript containing an adenosine insertion at the G2A6 site of TuMV and WYMV, respectively (Figure 4, Files S2 and S3). By contrast, the percentage of other transcript editing forms was below 1% (Figure 4). Combined with the mass spectrometry data, these results indicated that transcriptional slippage must be the expression mechanism of P3N-PIPO.
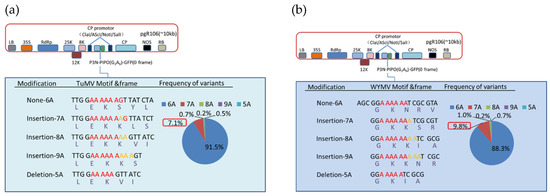
Figure 4.
Indel frequencies derived from deep sequencing at the G2A6 site of P3N-PIPO of TuMV and WYMV fused to GFP and expressed from a PVX vector in N. benthamiana. The P3N-PIPO-GFP constructs of TuMV (a) and WYMV (b) are depicted schematically. Details of the nucleotide sequences and resulting amino acid sequences of the translation products, as well as the RNA slippage frequencies detected for each modification are shown. Color codes in the pie charts refer to insertions (7A, 8A, 9A) or deletion (5A) compared to the wild type sequence (6A) of P3N-PIPO. The slippage frequency associated with a single adenosine insertion (7A) is highlighted in the red rectangular boxes.
3.4. Hexa-Adenosine at G2A6 Site Was Necessary for the Frameshift Expression of WYMV P3N-PIPO
To determine whether Hexa-adenosine was necessary for the frameshift expression of WYMV P3N-PIPO, two WYMV P3N-PIPO mutants were constructed, in which GGAAAAAA was mutated to GGACGCAA and CTAAAAAA, respectively. Thereafter, the frameshift expression of P3N-PIPO was monitored in inoculated N. benthamiana leaves under confocal microscopy. For wild-type P3N-PIPO (WT), GFP fluorescence was easily detected from 4 to 5 dpi in the infiltration area (Figure 5a). For P3N-PIPO M1, no GFP fluorescence was detected in the infiltration area at 5 and 10 dpi (Figure 5b), while for P3N-PIPO M2, GFP fluorescence was easily detected in the infiltration area at 5 dpi (Figure 5c). The behavior of P3N-PIPO M2 was similar to WT. These results indicated that Hexa-adenosine was important for the maintenance of P3N-PIPO frameshift expression.
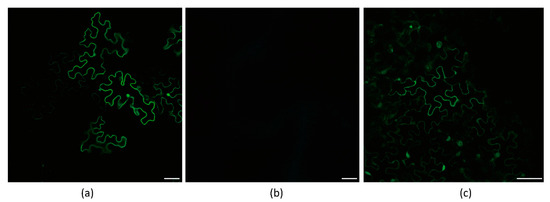
Figure 5.
Frameshift expression of WYMV P3N-PIPO mutants in N. benthamiana leaf cells under confocal microscopy. (a) PVX-WYMV P3N-PIPO-WT-GFP expressed in the epidermal cells treated eight days post-agroinfiltration; (b) PVX-P3N-PIPO-M1-GFP expressed in the epidermal cells treated eight days post-agroinfiltration; (c) PVX-P3NPIPO-M2-GFP expressed in the epidermal cells treated eight days post-agroinfiltration. Bars, 50 μm.
4. Discussion
The family Potyviridae comprises 10 genera and over 210 plant-infecting virus species, which can infect the most economically important crops in the world [26]. Most viruses in this family have single-stranded monopartite positive-sense RNA genomes, except for bipartite bymoviruses [2,26]. To date, studies on P3N-PIPO frameshift expression have been limited in the genus Potyvirus, and it remains unknown whether a similar frameshift expression of P3N-PIPO occurs in other genera in Potyviridae.
The in vivo frameshift expression of TuMV P3N-PIPO was proved by immunoblotting. Several reports have provided transcriptional evidence to support that P3N-PIPO is generated by polymerase slippage in a G2A6 conserved motif in potyviruses [2,19,27,28,29], and the single nucleotide insertion rate varies from 0.8 to 2.1% during the expression of P3N-PIPO [2,19]. Due to the extremely low expression level of P3N-PIPO in virus-infected plants, it is challenging to detect P3N-PIPO expression in vivo, let alone to identify the peptides spanning the slippery site.
Therefore, the PVX-based expression vector was used in this study to achieve an abundant P3N-PIPO expression for analysis, and TuMV P3N-PIPO(G2A6) was cloned into the PVX-based expression vector to test the validity of this approach. Our results indicated that the PVX-based expression vector was an excellent tool to study the expression mechanism of P3N-PIPO. In addition, peptides spanning the slippery site of P3N-PIPO were identified by mass spectrometry, which provided the first direct evidence for this frameshift expression.
To investigate the differences in the expression mechanism of P3N-PIPO between different genera in the family Potyviridae, P3N-PIPO(G2A6) of a bymovirus (WYMV) was cloned into the PVX-based expression vector using the same approach. Integrated proteome and transcriptome analyses confirmed that both P3N-PIPOs of TuMV and WYMV were expressed in the G2A6 conserved motif by means of transcriptional slippage.
In this study, the PVX-based vector system was successfully developed and proved to be an efficient and convenient tool for exploring the P3N-PIPO slippage efficiency. Furthermore, we provided the first consolidated evidence to support that P3N-PIPO of viruses in the genus Bymovirus showed frameshift expression in the G2A6 conserved motif. All in all, the PVX-based system developed in this study contributed to future research on the functional study of P3N-PIPO in Potyviridae.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13071247/s1, File S1: The peptides information of TuMV-P3N-PIPO(WT)-GFP and WYMV-P3N-PIPO(WT)-GFP identified by mass spectrometry analysis; File S2: The counts of indel sequences of PVX-TuMV-P3N-PIPO(WT)-GFP; File S3: The counts of indel sequences of PVX-WYMV-P3N-PIPO(WT)-GFP; Table S1: Primers used in this study.
Author Contributions
Conceptualization, C.Y., L.D. and J.C.; investigation, C.Y. and R.M., Y.L. and J.Y.; formal analysis S.M. and Z.Y.; writing—original draft preparation, C.Y.; writing—review & editing, J.L., F.Y., L.D. and J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No.31801703). This work was sponsored by the K.C. Wong Magna Fund in Ningbo University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Andrew J.M. Howden at the University of Dundee for his assistance with LC-MS/MS-based protein identification. We thank Peter Cock at the James Hutton Institute for advice on deep sequencing data analysis. We thank Yanzhen Tian for her assistance with confocal microscope work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maia, I.G.; Seron, K.; Haenni, A.-L.; Bernardi, F. Gene Expression from Viral RNA Genomes. Plant Mol. Biol. 1996, 32, 367–391. [Google Scholar] [CrossRef]
- Olspert, A.; Chung, B.Y.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional Slippage in the Positive-sense RNA Virus Family Potyviridae. EMBO Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Baranov, P.V.; Hammer, A.W.; Zhou, J.; Gesteland, R.F.; Atkins, J.F. Transcriptional Slippage in Bacteria: Distribution in Sequenced Genomes and Utilization in IS Element Gene Expression. Genome Biol. 2005, 6, R25. [Google Scholar] [CrossRef] [PubMed]
- Wagner, L.A.; Weiss, R.B.; Driscoll, R.; Dunn, D.S.; Gesteland, R.F. Transcriptional Slippage Occurs during Elongation at Runs of Adenine or Thymine in Escherichia Coli. Nucl. Acids Res. 1990, 18, 3529–3535. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Heath, L.S.; Turnbough, C.L. Regulation of PyrBI Operon Expression in Escherichia Coli by UTP-Sensitive Reiterative RNA Synthesis during Transcriptional Initiation. Genes Dev. 1994, 8, 2904–2912. [Google Scholar] [CrossRef][Green Version]
- Qi, F.; Turnbough, C.L., Jr. Regulation of codBA Operon Expression In Escherichia Coli by UTP-Dependent Reiterative Transcription and UTP-Sensitive Transcriptional Start Site Switching. J. Mol. Biol. 1995, 254, 552–565. [Google Scholar] [CrossRef]
- Sanchez, A.; Trappier, S.G.; Mahy, B.W.; Peters, C.J.; Nichol, S. The Virion Glycoproteins of Ebola Viruses Are Encoded in Two Reading Frames and Are Expressed through Transcriptional Editing. Proc. Natl. Acad. Sci. USA 1996, 93, 3602–3607. [Google Scholar] [CrossRef]
- Volchkov, V.E.; Becker, S.; Volchkova, V.A.; Ternovoj, V.A.; Kotov, A.N.; Netesov, S.V.; Klenk, H.-D. GP mRNA of Ebola Virus Is Edited by the Ebola Virus Polymerase and by T7 and Vaccinia Virus Polymerases1. Virology 1995, 214, 421–430. [Google Scholar] [CrossRef]
- Volchkova, V.A.; Dolnik, O.; Martinez, M.J.; Reynard, O.; Volchkov, V.E. Genomic RNA Editing and Its Impact on Ebola Virus Adaptation During Serial Passages in Cell Culture and Infection of Guinea Pigs. J. Infect. Dis. 2011, 204, S941–S946. [Google Scholar] [CrossRef]
- Ratinier, M.; Boulant, S.; Combet, C.; Targett-Adams, P.; McLauchlan, J.; Lavergne, J.P. Transcriptional Slippage Prompts Recoding in Alternate Reading Frames in the Hepatitis C Virus (HCV) Core Sequence from Strain HCV-1. J. Gen. Virol. 2008, 89, 1569–1578. [Google Scholar] [CrossRef]
- Hancock, J.M.; Chaleeprom, W.; Chaleeprom, W.; Dale, J.; Gibbs, A. Replication Slippage in the Evolution of Potyviruses. J. Gen. Virol. 1995, 76 Pt 12, 3229–3232. [Google Scholar] [CrossRef]
- Simón-Buela, L.; Osaba, L.; García, J.A.; López-Moya, J.J. Preservation of 5′-End Integrity of a Potyvirus Genomic RNA Is Not Dependent on Template Specificity. Virology 2000, 269, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.-W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An Overlapping Essential Gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the Trans-Frame Potyvirus Protein P3N-PIPO with Host Protein PCaP1 Facilitates Potyvirus Movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef]
- Wen, R.-H.; Hajimorad, M.R. Mutational Analysis of the Putative Pipo of Soybean Mosaic Virus Suggests Disruption of PIPO Protein Impedes Movement. Virology 2010, 400, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Cong, Q.-Q.; Li, X.-D.; Mou, A.-L.; Gao, R.; Liu, J.-L.; Tian, Y.-P. DEVELOPMENTALLY REGULATED PLASMA MEMBRANE PROTEIN of Nicotiana Benthamiana Contributes to Potyvirus Movement and Transports to Plasmodesmata via the Early Secretory Pathway and the Actomyosin System. Plant Physiol. 2015, 167, 394–410. [Google Scholar] [CrossRef]
- Chai, M.; Wu, X.; Liu, J.; Fang, Y.; Luan, Y.; Cui, X.; Zhou, X.; Wang, A.; Cheng, X. P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; Mingot, A.; San León, D.; Baulcombe, D.; López-Moya, J.J.; García, J.A. RNA Polymerase Slippage as a Mechanism for the Production of Frameshift Gene Products in Plant Viruses of the Potyviridae Family. J. Virol. 2015, 89, 6965–6967. [Google Scholar] [CrossRef]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato Virus X as a Vector for Gene Expression in Plants. Plant J. 1992, 2, 549–557. [Google Scholar]
- Dickmeis, C.; Fischer, R.; Commandeur, U. Potato Virus X-Based Expression Vectors Are Stabilized for Long-Term Production of Proteins and Larger Inserts. Biotechnol. J. 2014, 9, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. PGreen: A Versatile and Flexible Binary Ti Vector for Agrobacterium-Mediated Plant Transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Hayward, A.; Padmanabhan, M.; Dinesh-Kumar, S.P. Virus-Induced Gene Silencing in Nicotiana Benthamiana and Other Plant Species. Methods Mol. Biol. 2011, 678, 55–63. [Google Scholar] [PubMed]
- Granvogl, B.; Plöscher, M.; Eichacker, L.A. Sample Preparation by In-Gel Digestion for Mass Spectrometry-Based Proteomics. Anal. Bioanal. Chem. 2007, 389, 991–1002. [Google Scholar] [CrossRef]
- Thiede, B.; Lamer, S.; Mattow, J.; Siejak, F.; Dimmler, C.; Rudel, T.; Jungblut, P.R. Analysis of Missed Cleavage Sites, Tryptophan Oxidation and N-Terminal Pyroglutamylation after in-Gel Tryptic Digestion. Rapid Commun. Mass Spectrom. 2000, 14, 496–502. [Google Scholar] [CrossRef]
- Valli, A.; García, J.A.; López-Moya, J.J. Potyviruses (Potyviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 631–641. [Google Scholar]
- Untiveros, M.; Olspert, A.; Artola, K.; Firth, A.E.; Kreuze, J.F.; Valkonen, J.P.T. A Novel Sweet Potato Potyvirus Open Reading Frame (ORF) Is Expressed via Polymerase Slippage and Suppresses RNA Silencing. Mol. Plant Pathol. 2016, 17, 1111–1123. [Google Scholar] [CrossRef]
- Mingot, A.; Valli, A.; Rodamilans, B.; San León, D.; Baulcombe, D.C.; García, J.A.; López-Moya, J.J. The P1N-PISPO Trans-Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor. J. Virol. 2016, 90, 3543–3557. [Google Scholar] [CrossRef]
- White, K.A. The Polymerase Slips and PIPO Exists. EMBO Rep. 2015, 16, 885–886. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).