Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Plasmids and Viruses
2.3. Antibodies
2.4. Compounds
2.5. Compound Treatment during Transductions
2.6. Cytotoxicity Assay
2.7. Luciferase Assay
2.8. Immunoprecipitation Assay
2.9. Cell-Cell Fusion
2.10. Compound Treatment during IAV Infection
2.11. Western Blotting
2.12. RT-qPCR
2.13. Acidic Vacuole Staining
2.14. Immunofluorescence Staining
2.15. Statistics
3. Results
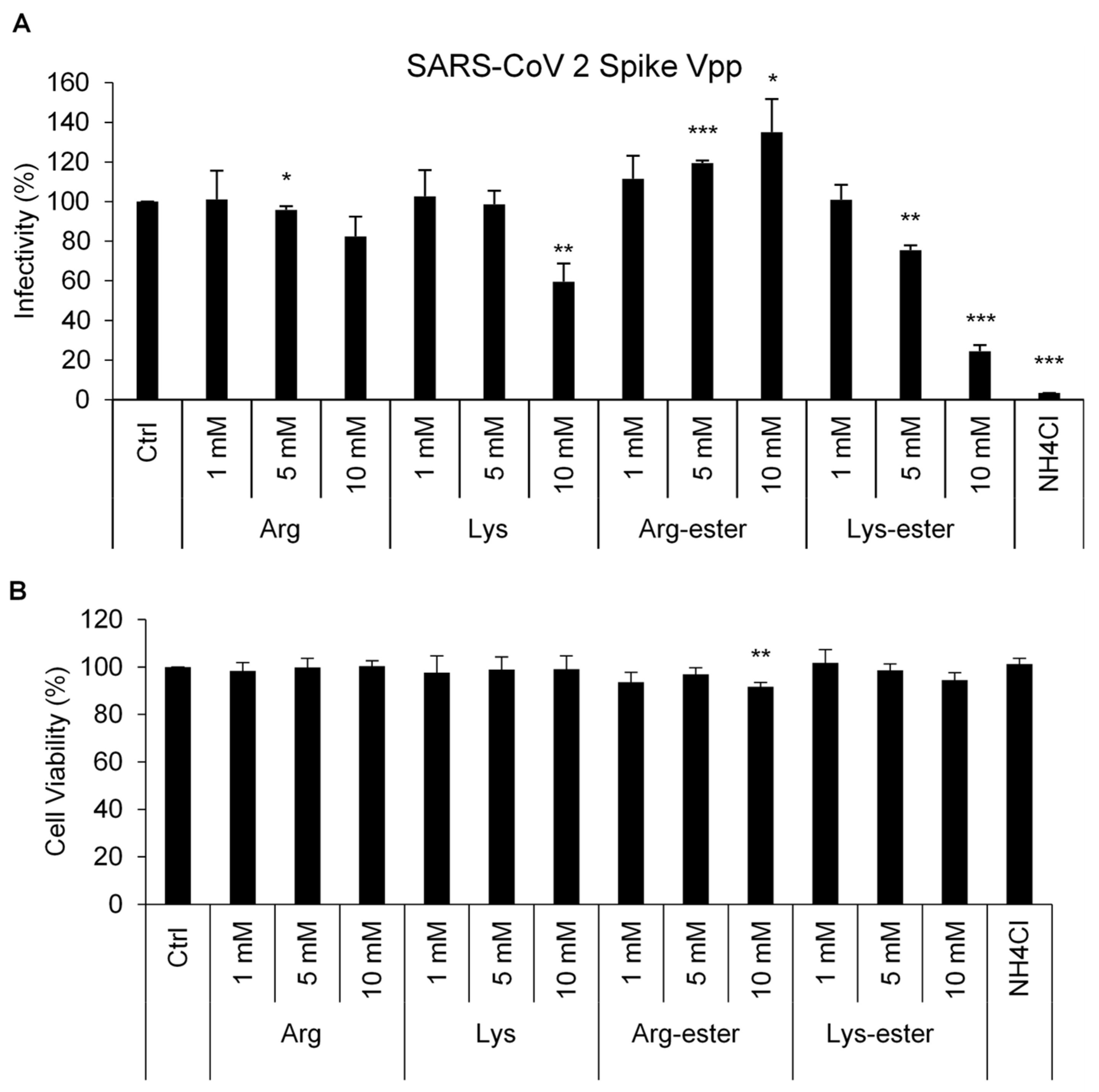
3.1. Lysine and Its Derivative Attenuate SARS-CoV-2 Vpp Infection
3.2. The Compounds Do Not Interfere the Interaction between Spike and ACE2
3.3. Lysine and Lys-Ester Inhibit IAV Replication
3.4. The Compounds Do Not Affect IAV Binding and Internalization
3.5. The Compounds Do Not Reduce Endosomal Acidification
3.6. Lysine and Lys-Ester Reduce Nuclear Distribution of IAV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Gayle, A.A.; Wilder-Smith, A.; Rocklöv, J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020, 27, taaa021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Liu, Y.; Zhao, M.; Zhuang, Q.; Xu, L.; He, Q. A comparison of COVID-19, SARS and MERS. PeerJ 2020, 8, e9725. [Google Scholar] [CrossRef]
- Kaur, N.; Singh, R.; Dar, Z.; Bijarnia, R.K.; Dhingra, N.; Kaur, T. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS-CoV2. Infect. Genet. Evol. 2021, 89, 104490. [Google Scholar] [CrossRef] [PubMed]
- Ngai, J.C.; Ko, F.W.; Ng, S.S.; To, K.-W.; Tong, M.; Hui, D.S. The long-term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology 2010, 15, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, R. As their numbers grow, COVID-19 “Long Haulers” stump experts. JAMA 2020, 324, 1381–1383. [Google Scholar] [CrossRef]
- Arbeitskreis Blut, U.B.B.K. Influenza virus. Transfus. Meds. Hemother. 2009, 36, 32–39. [Google Scholar] [CrossRef]
- Hashemi, S.A.; Safamanesh, S.; Ghasemzadeh-moghaddam, H.; Ghafouri, M.; Azimian, A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J. Med. Virol. 2021, 93, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhao, Y.; Dong, J.; Liang, S.; Guo, M.; Liu, X.; Wang, X.; Huang, Z.; Sun, X.; Zhang, Z.; et al. Co-infection of influenza A virus enhances SARS-CoV-2 infectivity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, A.J.; Lee, A.C.; Chan, J.F.; Liu, F.; Li, C.; Chen, Y.; Chu, H.; Lau, S.Y.; Wang, P.; Chan, C.C.; et al. Co-infection by severe acute respiratory syndrome coronavirus 2 and influenza A(H1N1)pdm09 virus enhances the severity of pneumonia in golden Syrian hamsters. Clin. Infect. Dis. 2020, 72, e978–e992. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Endocytosis of influenza viruses. Microbes Infect. 2004, 6, 929–936. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Sieben, C.; Ludwig, K.; Höfer, C.T.; Chiantia, S.; Herrmann, A.; Eghiaian, F.; Schaap, I.A.T. pH-Controlled two-step uncoating of influenza virus. Biophys. J. 2014, 106, 1447–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helenius, A. Virus entry: What has pH got to do with it? Nat. Cell Biol 2013, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Repnik, U.; Česen, M.H.; Turk, B. The endolysosomal system in cell death and survival. Cold Spring Harb. Perspect. Biol. 2013, 5, a008755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Chitalia, V.C.; Munawar, A.H. A painful lesson from the COVID-19 pandemic: The need for broad-spectrum, host-directed antivirals. J. Transl. Med. 2020, 18, 390. [Google Scholar] [CrossRef]
- Becht, H. Induction of an arginine-rich component during infection with influenza virus. J. Gen. Virol. 1969, 4, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Eaton, M.D.; Scala, A.R.; Low, I.E. Amino acid imbalance and incomplete viral replication. Arch. Für Die Gesamte Virusforsch. 1964, 14, 583–598. [Google Scholar] [CrossRef]
- Griffith, R.S.; DeLong, D.C.; Nelson, J.D. Relation of arginine-lysine antagonism to herpes simplex growth in tissue culture. Chemotherapy 1981, 27, 209–213. [Google Scholar] [CrossRef]
- Rossi, M.; Jacobs, B. Chapter 20—Herpes simplex virus. In Integrative Medicine (Fourth Edition); Rakel, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 191–197.e192. [Google Scholar] [CrossRef]
- Luiking, Y.C.; Deutz, N.E.P. Biomarkers of arginine and lysine excess. J. Nutr. 2007, 137, 1662S–1668S. [Google Scholar] [CrossRef] [Green Version]
- Mailoo, V.J.; Rampes, S. Lysine for herpes simplex prophylaxis: A review of the evidence. Integr. Med. (Encinitas) 2017, 16, 42–46. [Google Scholar]
- Griffith, R.S.; Norins, A.L.; Kagan, C. A multicentered study of lysine therapy in Herpes simplex infection. Dermatologica 1978, 156, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Griffith, R.S.; Walsh, D.E.; Myrmel, K.H.; Thompson, R.W.; Behforooz, A. Success of L-lysine therapy in frequently recurrent herpes simplex infection. Treatment and prophylaxis. Dermatologica 1987, 175, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Thein, D.J.; Hurt, W.C. Lysine as a prophylactic agent in the treatment of recurrent herpes simplex labialis. Oral Surg. Oral Med. Oral Pathol. 1984, 58, 659–666. [Google Scholar] [CrossRef]
- Postma, M.; Goedhart, J. PlotsOfData—A web app for visualizing data together with their summaries. PLoS Biol. 2019, 17, e3000202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Bari, M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharm. Res. Perspect 2017, 5, e00293. [Google Scholar] [CrossRef] [PubMed]
- Dabydeen, S.A.; Meneses, P.I. The role of NH4Cl and cysteine proteases in Human Papillomavirus type 16 infection. Virol. J. 2009, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matlin, K.S. Ammonium chloride slows transport of the influenza virus hemagglutinin but does not cause mis-sorting in a polarized epithelial cell line. J. Biol. Chem. 1986, 261, 15172–15178. [Google Scholar] [CrossRef]
- Oomens, A.G.P.; Wertz, G.W. The baculovirus GP64 protein mediates highly stable infectivity of a human respiratory syncytial virus lacking its homologous transmembrane glycoproteins. J. Virol. 2004, 78, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Su, W.-C.; Chen, Y.-C.; Tseng, C.-H.; Hsu, P.W.-C.; Tung, K.-F.; Jeng, K.-S.; Lai, M.M.C. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc. Natl. Acad. Sci. USA 2013, 110, 17516–17521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krolenko, S.A.; Adamyan, S.Y.; Belyaeva, T.N.; Mozhenok, T.P. Acridine orange accumulation in acid organelles of normal and vacuolated frog skeletal muscle fibres. Cell Biol. Int. 2006, 30, 933–939. [Google Scholar] [CrossRef]
- Bayer, N.; Schober, D.; Prchla, E.; Murphy, R.F.; Blaas, D.; Fuchs, R. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: Implications for viral uncoating and infection. J. Virol. 1998, 72, 9645–9655. [Google Scholar] [CrossRef] [Green Version]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef] [Green Version]
- Tankersley, R.W. Amino acid requirements of herpes simplex virus in human cells. J. Bacteriol. 1964, 87, 609–613. [Google Scholar] [CrossRef] [Green Version]
- Kagan, C.; Karlicki, B.; Chaihorsky, A.; Tal, R. Amino Acid L-Lysine SARS-CoV-2 (COVID-19) Prophylaxis; 2020; (Preprints). [Google Scholar] [CrossRef]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.-M.; Yang, W.-L.; Yang, F.-Y.; Zhang, L.; Huang, W.; Hou, W.; Fan, C.; Jin, R.; Feng, Y.; Wang, Y.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. medRxiv 2020. [Google Scholar] [CrossRef]
- Bertram, S.; Glowacka, I.; Steffen, I.; Kühl, A.; Pöhlmann, S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol 2010, 20, 298–310. [Google Scholar] [CrossRef]
- Civitelli, R.; Fedde, K.N.; Harter, J.; Halstead, L.R.; Gennari, C.; Avioli, L.V. Effect of L-lysine on cytosolic calcium homeostasis in cultured human normal fibroblasts. Calcif. Tissue Int. 1989, 45, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral calciomics: Interplays between Ca2+ and virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef]
- Liu, M.; Yang, Y.; Gu, C.; Yue, Y.; Wu, K.K.; Wu, J.; Zhu, Y. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J. 2007, 21, 1586–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartshorn, K.L.; Collamer, M.; Auerbach, M.; Myers, J.B.; Pavlotsky, N.; Tauber, A.I. Effects of influenza A virus on human neutrophil calcium metabolism. J. Immunol. 1988, 141, 1295–1301. [Google Scholar] [PubMed]
- Xing, Y.; Wen, Z.; Gao, W.; Lin, Z.; Zhong, J.; Jiu, Y. Multifaceted functions of host cell Caveolae/Caveolin-1 in virus infections. Viruses 2020, 12, 487. [Google Scholar] [CrossRef]
- Fecchi, K.; Anticoli, S.; Peruzzu, D.; Iessi, E.; Gagliardi, M.C.; Matarrese, P.; Ruggieri, A. Coronavirus Interplay with lipid rafts and autophagy unveils promising therapeutic targets. Front. Microbiol. 2020, 11, 1821. [Google Scholar] [CrossRef]
- Li, G.-M.; Li, Y.-G.; Yamate, M.; Li, S.-M.; Ikuta, K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007, 9, 96–102. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melano, I.; Kuo, L.-L.; Lo, Y.-C.; Sung, P.-W.; Tien, N.; Su, W.-C. Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection. Viruses 2021, 13, 1301. https://doi.org/10.3390/v13071301
Melano I, Kuo L-L, Lo Y-C, Sung P-W, Tien N, Su W-C. Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection. Viruses. 2021; 13(7):1301. https://doi.org/10.3390/v13071301
Chicago/Turabian StyleMelano, Ivonne, Li-Lan Kuo, Yan-Chung Lo, Po-Wei Sung, Ni Tien, and Wen-Chi Su. 2021. "Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection" Viruses 13, no. 7: 1301. https://doi.org/10.3390/v13071301
APA StyleMelano, I., Kuo, L.-L., Lo, Y.-C., Sung, P.-W., Tien, N., & Su, W.-C. (2021). Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection. Viruses, 13(7), 1301. https://doi.org/10.3390/v13071301





