1. Introduction
Site-specific cleavage of DNA with engineered nucleases forms the basis of gene editing techniques that are being developed to inactivate replication of hepatitis B virus (HBV) [
1]. Chronic infection with HBV is an important global health problem, and currently available therapies have modest curative efficacy [
2]. Fatal complicating cirrhosis and hepatocellular carcinoma are common amongst carriers of the virus, and account for approximately 887,000 HBV-related annual global deaths. Persistence of the essential HBV replication intermediate comprising covalently closed circular DNA (cccDNA) and minimal effects of licensed antivirals on this intermediate are the main reasons for current difficulties with eliminating HBV infection. Designer nucleases used to target cccDNA include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR) with CRISPR-associated (Cas) proteins [
1]. Targeted mutation is typically initiated by cleavage of a specific DNA sequence, which is then repaired by non-homologous end joining (NHEJ). With repeated cutting, error-prone DNA repair eventually leads to irreversible formation of replication-disabling insertions and deletions (indels) at the cleavage site.
CRISPR/Cas is now the most commonly applied gene editing tool, and the ease with which targeting nucleases may be generated is an important reason for popularity of the technology. These RNA-guided endonucleases have successfully been used to target DNA of HBV, and evidence indicates that cccDNA may be disabled in cells replicating the virus (reviewed in [
3]). However, a potential complication for therapy is the pervasive pre-existing immunity to the endonucleases derived from commensal
Streptococcus pyogenes or
Staphylococcus aureus [
4,
5]. Consequently, in vivo efficacy of candidate therapeutic gene editors may be compromised following systemic administration of sequences encoding anti-HBV CRISPR/Cas. Because TALENs and ZFNs are proteins derived from plant-infecting
Xanthomonas species or naturally occurring zinc finger proteins, pre-existing immunity is likely to be uncommon. To avoid immune attenuation, use of these gene editors may therefore be preferable to disable HBV. Comparisons between ZFNs and TALENs show that TALENs have advantages over ZFNs: unlike with ZFNs, DNA binding by individual TALEN monomers is not influenced by neighboring sequences, and TALENs have better specificity for their cognates than ZFNs [
6,
7].
Although gene editing technology has impressive potential, ensuring specificity of action is vital for therapeutic application. Off-target mutation caused by imprecise cleavage needs to be minimized to prevent potentially serious unintended consequences. Various approaches have been employed to improve precision of designer nucleases. In the case of CRISPR/Cas shortening of the guide sequence [
8], combining Cas nickases with two guides [
9,
10], inclusion of a hairpin structure in guide sequences [
11], and the recently described prime editing approach [
12] have all been used to achieve this goal. In the case of ZFNs and TALENs, the
FokI catalytic domain has been engineered in various ways to improve specificity. Slowing kinetics of target cleavage by
FokI, thereby selectively reducing action at low affinity off-target binding sites of ZFNs, has been successfully employed [
13]. Shortening the duration of action of gene editors may also diminish off-target effects, and this may be achieved by using mRNA as the coding nucleic acid [
14]. Modifying the
FokI nuclease domains to ensure formation of obligate heterodimers has also been utilized [
15,
16,
17]. The rationale for this approach is that juxtaposition of duplex-cleaving homodimers, comprising two left or two right subunits at an off-target site, is prevented. To avert generation of homodimers, researchers modified amino acid sequences at the interface between nuclease domains of these ZFNs, such that duplex-cleaving
FokI subunits were only active when heterodimers were assembled [
15,
16,
17]. In a similar vein, directed evolution has been employed to improve catalytic activity of the
FokI nuclease domain and yielded so-called Sharkey nuclease domains [
18]. We employed these approaches to improve specificity and activity of TALENs acting against HBV DNA by generating gene editors that require formation of obligate TALEN heterodimers to be active on their viral cognates. Evaluation in cultured cells and in vivo showed that the modified TALENs had similar activity to the first-generation counterparts, but with improved specificity to targets.
2. Materials and Methods
2.1. Plasmids
pCH-9/3091 [
19] is a replication-competent plasmid containing a greater-than-genome length HBV sequence. Transcription of the pCH-9/3091 plasmid is driven from the CMV promoter and yields a greater-than-genome length transcript that resembles the viral pgRNA, which subsequently initiates viral replication. pCI-neo eGFP [
20] and pCI-neo FLuc [
21] have been described before. Second-generation obligate heterodimeric TALENs and third-generation obligate heterodimeric TALENs with Sharkey mutations were derived from existing first-generation anti-HBV TALENs targeted against the
core and
surface ORFs of the viral genome [
22]. Each first-generation TALEN consists of left and right monomers comprising a DNA-binding TALE array fused to a first-generation
FokI nuclease domain with a hemagglutinin (HA) epitope and a nuclear localization signal (NLS) located at the N-terminus [
22]. The left and right monomers of the
core-targeting TALEN bind to nucleotides 2319–2337 and 2351–2369 of the HBV genome. The left and right monomers of the
surface-targeting TALEN bind to nucleotides 411–429 and 443–452 of the HBV genome [
22]. The first-generation anti-HBV TALENs exist within the pVAX plasmid backbone [
23] and are expressed from the CMV promoter. Second- and third-generation TALENs were generated by substituting the first-generation
FokI nuclease-encoding sequence in the pVAX plasmids with the second-generation obligate heterodimeric
FokI nuclease domain sequences or third-generation obligate heterodimeric
FokI nuclease domain sequence with Sharkey mutations, respectively.
2.2. Cell Culture
Huh7 cells were cultured in low-glucose DMEM (Thermo Scientific, Waltham, MA, USA), and HEK293 and HepG2.2.15 cells were maintained in high-glucose DMEM (Thermo Scientific, CA, USA). Growth medium was supplemented with penicillin (100,000 U/mL), streptomycin (100 μg/mL), and 10% Gibo™ FBS (Thermo Scientific, Waltham, MA, USA). Cells were maintained at 37 °C and 5% CO2 in a humidified incubator.
2.3. Immunofluorescence Detection of Anti-HBV TALEN Expression
Twenty-four hours before transfection, Huh7 cells were seeded in a 96-well plate at a density of 50% per well. Lipofectamine® 3000 (Thermo Scientific, Waltham, MA, USA) was used to transfect 100 ng of each TALEN monomer-expressing plasmid. pCI-neo eGFP was transfected separately to verify success of transfection. Forty-eight hours after transfection, TALEN expression was assessed by immunofluorescence detection of the HA epitope using an anti-HA primary antibody (Sigma-Aldrich, St. Louis, MO, USA) and Alexa Fluor 448-labeled secondary antibody (Thermo Scientific, Waltham, MA, USA). Fluorescence was detected using the Axiovert 100M fluorescence microscope (Zeiss, Oberkochen, Germany).
2.4. Assessment of HBV Silencing in Cultured Cells by ELISA
Huh7 cells were seeded in a 6-well plate at a cell density of 50% and transfected one day later. Using Lipofectamine® 3000, cells were transfected with 1 µg of each left TALEN monomer-expressing plasmid together with 1 µg of its cognate right TALEN monomer-expressing plasmid, 300 ng of pCH-9/3091, and 200 ng of pCI-neo eGFP. As a mock, 2 µg of pUC118 (Addgene, Watertown, MA, USA) was co-transfected with pCH-9/3091 and pCI-neo eGFP. Forty-eight hours post-transfection, successful transfection was determined by visualizing GFP expression and HBsAg secretion. GFP expression was detected using fluorescence microscopy and HBsAg was assayed using ELISA with the Monolisa™ HBsAg ULTRA kit (Bio-Rad, Hercules, CA, USA).
2.5. On-Target Cleavage by Anti-HBV TALENs Using the SURVEYOR Assay
HepG2.2.15 cells were seeded in 6-well plates and transfected the following day with 200 ng pCI-neo eGFP and 1 µg of each of the left and right TALEN monomer-encoding plasmids using Lipofectamine
® 3000. As a control, 2 µg of pUC118 was transfected in place of the TALEN-expressing plasmids. Spent medium was replaced three days post-transfection. After five days, half the cells were harvested and the other half re-seeded. Transfection and re-seeding were repeated for an additional 2 cycles. After the final transfection, supernatants were collected for HBsAg ELISA, and cells were harvested and used to assess targeted cleavage. Total DNA was extracted from HepG2.2.15 cells as previously described [
22]. Sequences comprising 520 bp and spanning ≈260 bp upstream and ≈260 bp downstream of the predicted target sites for the C and S TALENs were amplified under standard PCR conditions. The following primer sets were used: Core forward 5′-GAA CTA ATG ACT CTA GCT ACC T-3′, Core reverse 5′-CCT ACA AAC TGT TCA CAT TT-3′; Surface forward 5′-CCT AGG ACC CCT TTC TCG TGT-3′, and Surface reverse 5′-ACT GAG CCA GGA GAA ACG GG-3′. Three hundred nanograms of PCR products were subjected to heteroduplex formation by denaturation at 95 °C followed by cooling to 35 °C at a ramp rate of −0.1 °C/s, then holding at 35 °C for 2 min. At this point, PCR products were incubated on ice for 5 min, followed by the addition of 1 µL of CelI enzyme and 2 µL of 10× NEB buffer 2 (New England Biolabs, Ipswich, MA, USA). Samples were held at 4 °C for 10 min, followed by heating and incubation at 37 °C for 25 min. Cleaved products were resolved using agarose gel electrophoresis, and ImageJ software was used to measure targeted disruption as previously described [
24]. As a positive control for the Surveyor assay, heteroduplexes were formed by the PCR amplification of first-generation and mutant HBx sequences [
25]. First-generation and mutant amplicons were mixed at equimolar amounts and denatured and annealed to form heteroduplexes.
2.6. Assessment of Cell Viability by MTT Assay
HEK293 cells were seeded in a 96-well plate at a density of 30% six hours prior to transfection. Polyethylenimine (PEI) (0.1 mg/mL) was used to co-transfect 15 ng of pCH-9/3091, 15 ng of pCI-neo eGFP, and 85 ng of the left and right TALEN monomer-expressing plasmids. Cells transfected with 170 ng of pUC118 served as the mock transfection control. Cells treated with 50% dimethyl sulfoxide (DMSO) were used as a positive control, and untreated cells served as the negative control. Cell viability was assessed 48 h after transfection. Twenty microliters of 5 mg/mL MTT, made up in PBS, was added to each well and incubated at 37 °C for 1 h. Culture medium was subsequently removed, 200 µL of DMSO added and the cells incubated for a further 5 min. The metabolism of MTT to form blue formazan was determined by measuring the optical densities at 570 nm and 655 nm using an iMARK™ Microplate reader (Bio-Rad, Hercules, CA, USA).
2.7. Animal Studies
Anti-HBV TALEN efficacy was assessed using the murine hydrodynamic injection (HDI) model of HBV replication. All experiments on animals were conducted in accordance with protocols approved by the University of the Witwatersrand Animal Research Ethics Committee. HDI was performed on 5-week-old female NMRI mice, weighing between 20 and 30 g. The injectate comprised a plasmid DNA-containing saline solution equal to 10% of the body weight of each mouse (final volume of 2–3 mL). The DNA/saline solutions contained 5 µg HBV target DNA (pCH-9/3091), 5 µg pCI-neo FLuc, and either 20 µg of pUC118 or 10 µg of each corresponding left and right TALEN-expressing plasmids. All plasmids were prepared using the Endo-Free Plasmid Maxi kit (Qiagen, Hilden, Germany). To confirm hepatic delivery of the plasmids, 3 days post-injection mice were injected intraperitoneally with 150 mg/kg of D-luciferin (PerkinElmer, Inc., Waltham, MA, USA) and bioluminescence imaging carried out using an IVIS Kinetic In Vivo Optical Imaging System (PerkinElmer, Inc., Waltham, MA, USA). Blood was collected from mice by retro-orbital puncture on days 3 and 5 post-injection, and the serum was then diluted in an equal volume of saline. One hundred microliters were used to measure serum HBsAg concentrations using the Monolisa™ HBsAg ULTRA kit. Serum ALT levels were quantified using a kinetic assay with an automated photometric analyzer (Roche Applied Science, Penzberg, Germany). Mice were euthanized on day 5 by CO2 exposure, and livers were then immediately harvested. To assess targeted cleavage in murine samples, we dissected and mechanically homogenized 25 mg of liver. Total DNA was extracted from liver homogenates using the QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany). On-target cleavage was assessed using the Surveyor assay as described earlier.
2.8. Quantification of Circulating VPEs and Gene Expression
To quantify viral particle equivalents (VPEs), we extracted total DNA from 50 µL of diluted serum using the QIAamp® DNA Mini Kit. Circulating VPEs from experimental and control mice were measured by qPCR using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The Acrometrix HBV Panel (Thermo Scientific, Waltham, WA, USA) was used as a standard for quantitation. DNA samples were subjected to real-time PCR using 2× FastStart Essential DNA Green Master (Roche Applied Science, Penzberg, Germany) with the following primers: HBVs F 5′-TGC ACC TGT ATT CCC ATC-3′ and HBVs R 5′-CTG AAA GCC AAA CAG TGG-3′. To assess intrahepatic HBV gene expression, we quantified viral RNA levels by RT-qPCR. Total cellular RNA was extracted from liver homogenates using the TRIzol® Reagent (Thermo Scientific, Waltham, WA, USA) and reverse transcribed using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). The cDNA samples were subjected to real-time PCR using 2× FastStart Essential DNA Green Master. Viral surface and pregenomic RNA were amplified using the HBVs F and R primers, and a second primer set (BCP F 5-′ACC ACC AAA TGC CCC TAT-3′ and BCP R 5′-TTC TGC GAG GCG GCG A-3′) was used to amplify pregenomic RNA selectively. Murine GAPDH was amplified with the mGAPDH F 5′-TTC ACC ACC ATG GAG AAG GC-3′ and mGAPDH R 5′-GGC ATG GAC TGT GGT CAT GA-3′ primers to relativize viral RNA levels.
2.9. Assessment of on- and off-Target Mutagenesis by Next Generation Sequencing
Potential off-target sites within the host genome were predicted for the S and C TALENs using PROGNOS software [
26], and the top four off-target sites were selected for further analysis. Four mice were chosen from each group and total DNA was extracted from homogenized liver samples using the QIAamp
® DNA Blood Mini Kit. Sequences flanking the on-target site and potential off-target sites were amplified using the KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA) with primer sets listed in
Table S1. The primers were designed to amplify a 250–300 bp region flanking the on-target site and each of the four off-target sites. The PCR amplicons were column-purified using the MinElute Gel Extraction kit and pooled on the basis of group and target amplicon. Primer sets contained Multiplex IDentifiers (MIDs) to allow discrimination of the different mice from each other. Each amplicon had a 10 bp MID flanking the different regions of interest. Samples were sequenced using the HiSeq 2500 System (Illumina, San Diego, CA, USA). The generated paired-end reads were merged using Flash (
https://ccb.jhu.edu/software/FLASH/; October 2020 to June 2021) and demultiplexed according to the MIDs that were used for each sample (
https://github.com/najoshi/sabre, accessed on 30 June 2021). Merged reads were further analyzed using the command line version of CRISPResso2 [
27] with a window size of 30, substitutions were ignored.
2.10. Data Analysis
Data were presented as mean ± SEM. Two-tailed Student’s t-tests were performed using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA, USA) for the comparison between two groups. A value of p < 0.05 (*) was considered statistically significant.
4. Discussion
ZFNs and TALENs are designed as left and right monomers that come together at the intended DNA target sequences, allowing their
FokI nuclease domains to dimerize and create a double-stranded break. However, left/left or right/right homodimers may also assemble allowing functional
FokI dimerization and cleavage at these unintended sites. To limit this possibility, second- and third-generation
FokI nuclease domains were identified that are only functional when left/right heterodimerization occurs [
15,
16,
17]. Q486E, together with I499A and E490K, together with I538V, for example, yielded ZFN monomers that function poorly as homodimers but very efficiently as heterodimers [
16]. A subsequent study identified additional modifications to the
FokI nuclease domain, such as ELD (Q486E, I499L, N496D) and KKR (E490K, I538K, H537R) mutations, which yielded improved obligate heterodimeric ZFNs [
17]. Modifications that enhance the catalytic activity of the
FokI nuclease domain and improve efficacy of ZFNs have also been identified [
18]. These so-called Sharkey mutations (S418P and K441E) may also reduce off-target mutagenesis of nucleases as lower dosages would be required to produce a therapeutic effect.
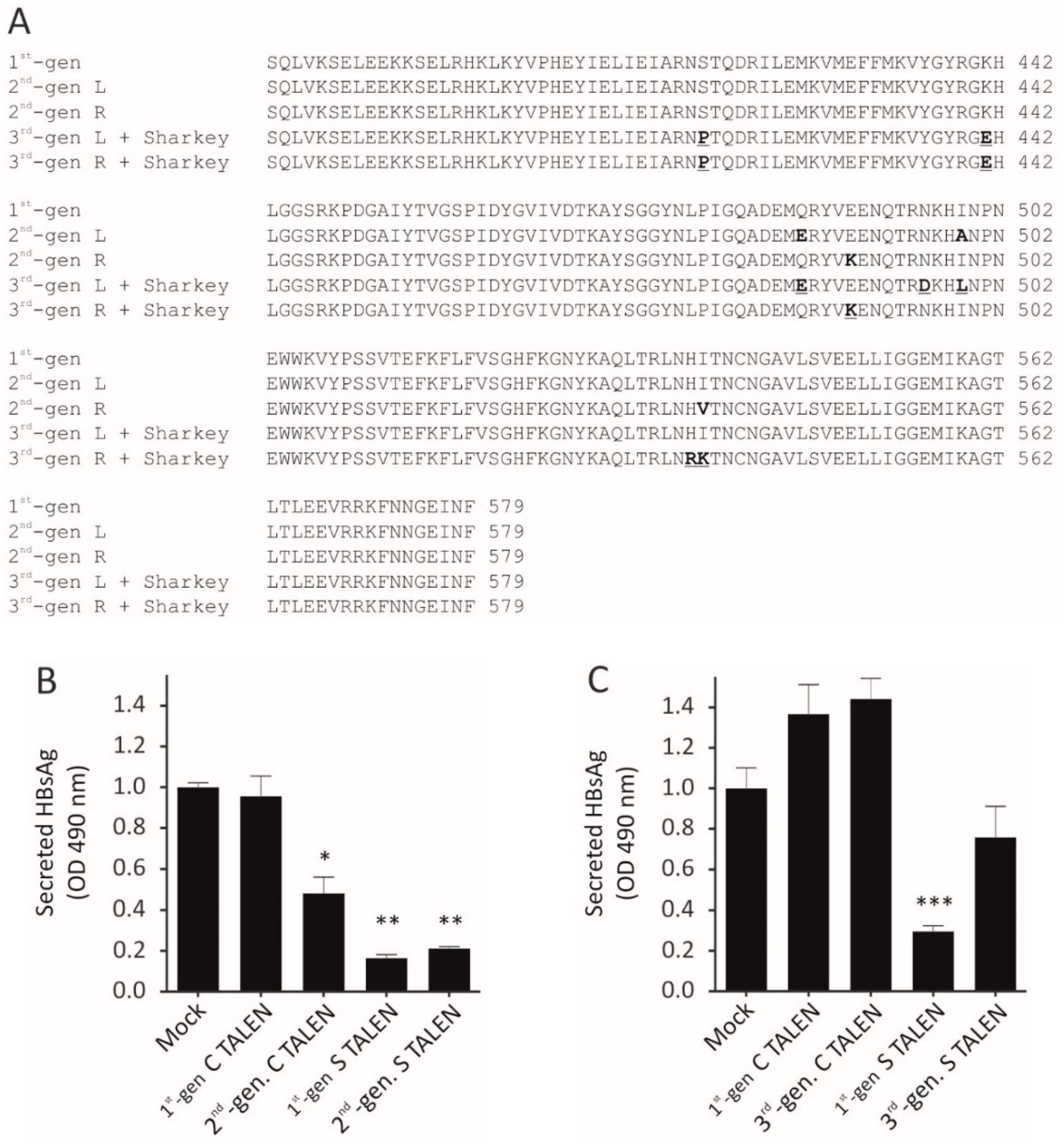
Here, we evaluated the use of second-generation TALENs (Q486E, I499A, E490K, and I538V obligate heterodimers) as well as third-generation TALENs (ELD and KKR obligate heterodimers) with Sharkey mutations for use against HBV. In general, the efficacy of the second-generation obligate heterodimers were on par with that of the original first-generation anti-HBV TALENs. However, although similar levels of suppression of viral replication by the second-generation obligate heterodimeric TALEN targeted to the
core ORF were observed, lower target disruption was observed in vivo. In contrast the third-generation TALENs containing Sharkey mutations exhibited reduced silencing activity against HBV. Similar results have been reported before [
28,
29]. This suggests that incorporating Sharkey mutations into the TALEN architecture, and more specifically into third-generation
FokI nuclease domains, is deleterious to silencing activity [
28,
29]. The Sharkey mutations were generated by directed evolution of ZFNs and possibly provides the reason for reduced silencing activity within TALENs.
Unexpectedly, the anti-HBV TALENs reduced viral mRNA levels. The likely explanation for this observation is that, in addition to their nuclease function, the TALENs were capable of suppressing viral DNA at the transcriptional level. Although the TALENs described here do not contain transcription inhibitory domains, it has been demonstrated that ZFPs comprising only a DNA-binding domain were capable of suppressing duck hepatitis B virus (DHBV) transcription [
30]. The ZFPs were targeted to the enhancer region of DHBV, which controls core and small surface protein expression, which were suppressed as a result. Unexpectedly, production of DHBV large surface protein, which is not under the control of the enhancer region, was also inhibited. Steric hindrance of RNA polymerase by the ZFPs that prevented transcription of the large surface protein gene was postulated as the mechanism for the observed indirect suppression. Another study reported inhibition of HBsAg secretion by a TALEN targeted against the
polymerase ORF of HBV and speculated that the mechanism might be mediated by transcriptional interference [
22]. Transcriptional repression by the anti-HBV TALENs may explain the results reported here. The lack of any elevation in serum ALT levels suggests the suppression is not as a result of non-specific effects.
Characterization of off-target mutagenesis in vivo using NGS suggested that second-generation obligate heterodimeric TALENs exhibit improved specificity. Extensive mutation of an intronic region of the phenylalanine hydroxylase gene by the first-generation C TALEN was observed, whereas the obligate heterodimeric C TALEN produced fewer mutations at this site. This observation is supported by the fact that this off-target site is predicted to be targeted by a right/right homodimer. Although lower, targeted mutagenesis induced by the obligate heterodimeric C TALEN was nevertheless substantive, suggesting that specificity of these nucleases may still require improvement.
Studies evaluating therapeutic interventions against HBV are plagued by the poor models of chronic hepatitis B. Cell culture models of viral replication, such as transient transfection of liver-derived cells or stable HBV cell lines used here, do not recapitulate all aspects of chronic infection. Cultured cells, in particular, do not model viral integration or the existence of HBV quasi-species as seen in chronic carriers. The HDI model of HBV replication used in this study, too, does not fully model the natural infection process. Of note, the mouse hepatocyte does not support cccDNA formation and as a consequence the ability of the TALENs to target this viral intermediate cannot be determined in vivo. Furthermore, HDI necessitates the co-administration of the replication-competent HBV plasmid with the TALEN-encoding plasmids, which does not model post-exposure intervention.
The ability to directly target and inactivate cccDNA makes the use of engineered nucleases, such as TALENs, a worthy avenue to be explored as a therapeutic modality for chronic HBV infection. TALEN and Cas9 function has been shown to be limited by heterochromatin [
31]; more recent data suggest that TALENs fare better than the Cas9 nuclease at navigating compact DNA [
32]. This is important in HBV therapy as the cccDNA has been shown to exist in a heterochromatic state [
33,
34]. While TALENs are obstructed by heterochromatin, activity is not completely inhibited and the nuclease is capable of navigating within heterochromatin, more so than the Cas9 nuclease, which has to separate double stranded DNA before interrogating the target site [
32]. Effective targeting of the viral DNA relies on binding of the TALENs to the target sequences, which may be disrupted by escape mutations. Analysis of HBV sequencing data identified limited variability in the target sites of the TALENs described here [
22]. Furthermore, targeting multiple sites within the HBV genome simultaneously will be necessary to limit viral escape. Characterization of off-target effects and development of an efficient delivery vehicle for TALEN-expressing sequences remain crucial to the eventual clinical translation of this technology. Advances in sequencing technology have yielded a wealth of information and will play an important role in identifying off-target disruption by engineered nucleases. TALENs, in particular, face the challenge of delivering two very large transgenes to the same cell to be effective. While viral vectors have been explored extensively for this purpose, the potential for recombination of repeat sequences limits their utility. Use of in vitro-transcribed mRNA with synthetic vectors may offer advantages of safety and facile large-scale manufacturing capability. TALEN technology is well-placed to fill a significant gap in anti-HBV therapeutics.
The management of HBV is plagued by poor vaccine coverage and ineffective treatment options, and as a consequence, disease burden globally, especially in resource-poor settings, remains high. Acute infections are estimated to be responsible for close to 100,000 deaths annually, but mortality from chronic hepatitis B-associated complications far exceed this number [
35]. There is therefore an urgent need for curative therapies to combat chronic HBV infection. Persistence of the infection stems from the viral cccDNA, which is established as a stable episomal minichromosome during infection. Effecting a functional cure of chronic hepatitis B, involving complete suppression of cccDNA activity, is increasingly recognized as a goal capable of being achieved over that of a sterilizing cure, which necessitates the complete removal of all viral reservoirs. Engineered nucleases have the potential to directly target cccDNA and induce disruptive mutations to permanently inactivate this viral intermediate. For the eventual application of engineered nucleases in a clinical setting, undesired gene disruption at unintended target sites needs to be eliminated.