Setting the Terms for Zoonotic Diseases: Effective Communication for Research, Conservation, and Public Policy
Abstract
1. Introduction
- Incorrect or overly broad use of terms—words in which the sender is unaware of the accepted definition of a term in the field(s) that coined it and so uses the term erroneously, or without the level of evidence that would support that use, e.g., bats as the reservoir of MERS-CoV or SARS-CoV-2. This perpetuates misinformation and misunderstanding in audiences, whether they are other researchers or the general public.
- Terms that have unstable usage within a discipline (e.g., bat origin), or different usages among disciplines (e.g., endemic, vector), leading to confusion or misunderstanding in audiences.
- Terms that are used correctly but spark incorrect inferences about biological processes or significance in the audience (e.g., spillover, novel).
- Incorrect inference from the evidence presented. This may be due to the fact that the audience (and occasionally the messenger) is unfamiliar with the methodologies generating the evidence (e.g., serological evidence, phylogenetic evidence).
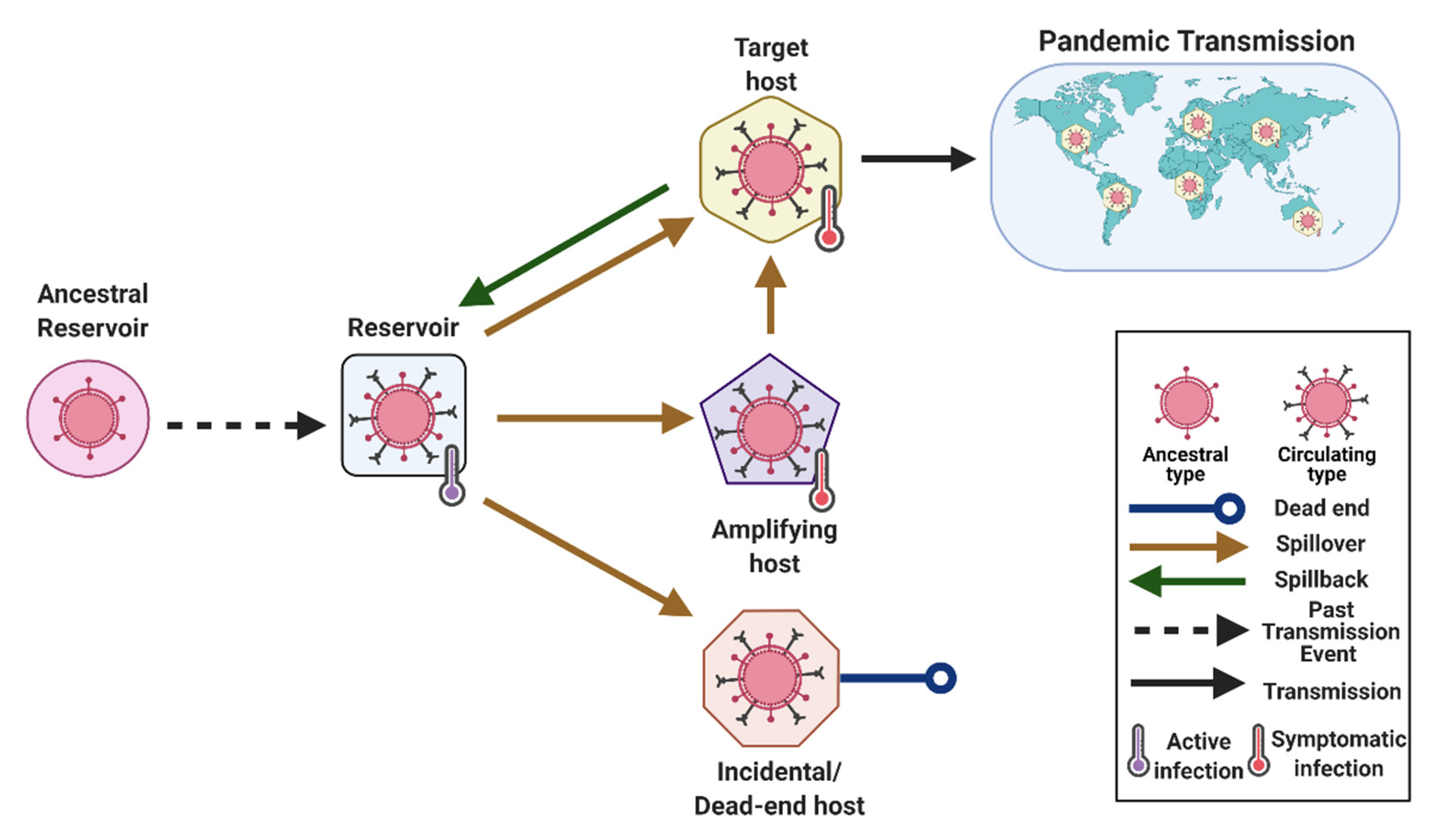
2. Glossary of Terms
- a.
- Entities
- (i.)
- Pathogen (Type 1)
- (ii.)
- Reservoir or Reservoir Host (Type 1)
- (iii.)
- Intermediate host (Type 2, Type 1)
- (iv.)
- Vector (Type 2)
- b.
- Processes and Events
- (i.)
- Spillover (Type 3)
- (ii.)
- Spillback (Type 1)
- (iii.)
- Mutation (Type 3)
- c.
- Descriptors:
- (i.)
- Novel/New (Type 3)
- (ii.)
- Endemic (Type 2)
- (iii.)
- Bat origin (Type 2, Type 3)
- (iv.)
- Bats (Type 1)
- d.
- Methodologies:
- (i.)
- Serology (Type 4)
- (ii.)
- Phylogeny (Type 4)
3. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kading, R.C.; Kingston, T. Common Ground: The Foundation of Interdisciplinary Research on Bat Disease Emergence. PLoS Biol. 2020, 18, e3000947. [Google Scholar] [CrossRef]
- Burgin, C.J.; Colella, J.P.; Kahn, P.L.; Upham, N.S. How Many Species of Mammals Are There? J. Mammal. 2018, 99, 1–14. [Google Scholar] [CrossRef]
- Anderson, D.E.; Islam, A.; Crameri, G.; Todd, S.; Islam, A.; Khan, S.U.; Foord, A.; Rahman, M.Z.; Mendenhall, I.H.; Luby, S.P.; et al. Isolation and Full-Genome Characterization of Nipah Viruses from Bats, Bangladesh. Emerg. Infect. Dis. 2019, 25, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Leendertz, S.A.J.; Gogarten, J.F.; Düx, A.; Calvignac-Spencer, S.; Leendertz, F.H. Assessing the Evidence Supporting Fruit Bats as the Primary Reservoirs for Ebola Viruses. EcoHealth 2015, 13, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Leendertz, S.A.J. Testing New Hypotheses Regarding Ebolavirus Reservoirs. Viruses 2016, 8, 30. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Lu, M.; Wang, X.; Ye, H.; Wang, H.; Qiu, S.; Zhang, H.; Liu, Y.; Luo, J.; Feng, J. Does Public Fear That Bats Spread COVID-19 Jeopardize Bat Conservation? Biol. Conserv. 2021, 108952. [Google Scholar] [CrossRef] [PubMed]
- López-Baucells, A.; Rocha, R.; Fernández-Llamazares, Á. When Bats Go Viral: Negative Framings in Virological Research Imperil Bat Conservation. Mammal Rev. 2018, 48, 62–66. [Google Scholar] [CrossRef]
- MacFarlane, D.; Rocha, R. Guidelines for Communicating about Bats to Prevent Persecution in the Time of COVID-19. Biol. Conserv. 2020, 248, 108650. [Google Scholar] [CrossRef]
- Sasse, B.; Gramza, A. Influence of the COVID-19 Pandemic on Public Attitudes toward Bats in Arkansas and Implications for Bat Management. Hum. Dimens. Wildl. 2021. [Google Scholar] [CrossRef]
- Littlejohn, S.W.; Foss, K.A. Theories of Human Communication, 10th ed.; Waveland Press: Long Grove, IL, USA, 2010; ISBN 978-1-4786-0939-1. [Google Scholar]
- Margolis, L.; Esch, G.W.; Holmes, J.C.; Kuris, A.M.; Schad, G.A. The Use of Ecological Terms in Parasitology (Report of an Ad Hoc Committee of the American Society of Parasitologists). J. Parasitol. 1982, 68, 131–133. [Google Scholar] [CrossRef]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology Meets Ecology on Its Own Terms: Margolis et al. Revisited. J. Parasitol. 1997, 83, 575. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L. Host-Pathogen Interactions: Redefining the Basic Concepts of Virulence and Pathogenicity. Infect. Immun. 1999, 67, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.-A. What Is a Pathogen? Ann. Med. 2002, 34, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Pirofski, L.; Casadevall, A. Q&A: What Is a Pathogen? A Question That Begs the Point. BMC Biol. 2012, 10, 6. [Google Scholar] [CrossRef]
- Olival, K.J.; Hayman, D.T.S. Filoviruses in Bats: Current Knowledge and Future Directions. Viruses 2014, 6, 1759–1788. [Google Scholar] [CrossRef]
- Curto, P.; Santa, C.; Allen, P.; Manadas, B.; Simões, I.; Martinez, J.J. A Pathogen and a Non-Pathogen Spotted Fever Group Rickettsia Trigger Differential Proteome Signatures in Macrophages. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G. The Disease Triangle: Pathogens, the Environment and Society. Nat. Rev. Microbiol. 2007, 5, 152–156. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.P.; Procop, G.W.; Wilson, M. Infectious Disease Pathology. Clin. Infect. Dis. 2001, 32, 1589–1601. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Viral Subversion of the Host Protein Synthesis Machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Capucci, L.; Fusi, P.; Lavazza, A.; Pacciarini, M.L.; Rossi, C. Detection and Preliminary Characterization of a New Rabbit Calicivirus Related to Rabbit Hemorrhagic Disease Virus but Nonpathogenic. J. Virol. 1996, 70, 8614–8623. [Google Scholar] [CrossRef]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P.J.Y. Characterization of Pathogenic and Non-Pathogenic African Swine Fever Virus Isolates from Ornithodoros erraticus Inhabiting Pig Premises in Portugal. J. Gen. Virol. 2004, 85, 2177–2187. [Google Scholar] [CrossRef]
- Marsh, G.A.; de Jong, C.; Barr, J.A.; Tachedjian, M.; Smith, C.; Middleton, D.; Yu, M.; Todd, S.; Foord, A.J.; Haring, V.; et al. Cedar Virus: A Novel Henipavirus Isolated from Australian Bats. PLoS Pathog. 2012, 8, e1002836. [Google Scholar] [CrossRef]
- Fredericks, D.N.; Relman, D.A. Sequence-Based Identification of Microbial Pathogens: A Reconsideration of Koch’s Postulates. Clin. Microbiol. Rev. 1996, 9, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Hul, V.; Delaune, D.; Karlsson, E.A.; Hassanin, A.; Tey, P.O.; Baidaliuk, A.; Gámbaro, F.; Tu, V.T.; Keatts, L.; Mazet, J.; et al. A Novel SARS-CoV-2 Related Coronavirus in Bats from Cambodia. bioRxiv 2021. [Google Scholar] [CrossRef]
- Culbertson, Alix COVID-19: Bats Living in Cambodia in 2010 Carried “nearly Identical” Pathogen to COVID-19 Virus, Scientists Discover. Sky News, 2021. Available online: https://news.sky.com/story/covid-19-bats-living-in-cambodia-in-2010-carried-nearly-identical-pathogen-to-covid-19-virus-scientists-discover-12201222 (accessed on 20 February 2021).
- Goldstein, T.; Anthony, S.J.; Gbakima, A.; Bird, B.H.; Bangura, J.; Tremeau-Bravard, A.; Belaganahalli, M.N.; Wells, H.L.; Dhanota, J.K.; Liang, E.; et al. The Discovery of Bombali Virus Adds Further Support for Bats as Hosts of Ebolaviruses. Nat. Microbiol. 2018, 3, 1084–1089. [Google Scholar] [CrossRef]
- Ashford, R.W. What It Takes to Be a Reservoir Host. Belg. J. Zool. 1997, 127, 85–90. [Google Scholar]
- Ashford, R.W. When Is a Reservoir Not a Reservoir? Emerg. Infect. Dis. 2003, 9, 1495–1496. [Google Scholar] [CrossRef]
- Hubálek, Z. Emerging Human Infectious Diseases: Anthroponoses, Zoonoses, and Sapronoses. Emerg. Infect. Dis. 2003, 9, 403–404. [Google Scholar] [CrossRef]
- Haydon, D.T.; Cleaveland, S.; Taylor, L.H.; Laurenson, M. Karen Identifying Reservoirs of Infection: A Conceptual and Practical Challenge. Emerg. Infect. Dis. 2002, 8, 1468–1473. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats Host Major Mammalian Paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef]
- Vial, F.; Cleaveland, S.; Rasmussen, G.; Haydon, D.T. Development of Vaccination Strategies for the Management of Rabies in African Wild Dogs. Biol. Conserv. 2006, 131, 180–192. [Google Scholar] [CrossRef]
- Lembo, T.; Hampson, K.; Haydon, D.T.; Craft, M.; Dobson, A.; Dushoff, J.; Ernest, E.; Hoare, R.; Kaare, M.; Mlengeya, T.; et al. Exploring Reservoir Dynamics: A Case Study of Rabies in the Serengeti Ecosystem. J. Appl. Ecol. 2008, 45, 1246–1257. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Mayer, A.E.; Niezgoda, M.; Markotter, W.; Agwanda, B.; Breiman, R.F.; Rupprecht, C.E. Shimoni Bat Virus, a New Representative of the Lyssavirus Genus. Virus Res. 2010, 149, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, I.V.; Turmelle, A.S.; Agwanda, B.; Markotter, W.; Niezgoda, M.; Breiman, R.F.; Rupprecht, C.E. Commerson’s Leaf-Nosed Bat (Hipposideros commersoni) Is the Likely Reservoir of Shimoni Bat Virus. Vector-Borne Zoonotic Dis. 2011, 11, 1465–1470. [Google Scholar] [CrossRef]
- Foley, N.M.; Goodman, S.M.; Whelan, C.V.; Puechmaille, S.J.; Teeling, E. Towards Navigating the Minotaur’s Labyrinth: Cryptic Diversity and Taxonomic Revision within the Speciose Genus Hipposideros (Hipposideridae). Acta Chiropterologica 2017, 19, 1–18. [Google Scholar] [CrossRef]
- Müller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.M.; Bosch, B.-J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS Coronavirus Neutralizing Antibodies in Camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, V.S.; Zaki, A.M.; Osterhaus, A.D.M.E.; Haagmans, B.L.; Gorbalenya, A.E.; Snijder, E.J.; et al. Genomic Characterization of a Newly Discovered Coronavirus Associated with Acute Respiratory Distress Syndrome in Humans. mBio 2012, 3. [Google Scholar] [CrossRef]
- Reusken, C.B.; Haagmans, B.L.; Müller, M.A.; Gutierrez, C.; Godeke, G.-J.; Meyer, B.; Muth, D.; Raj, V.S.; Vries, L.S.-D.; Corman, V.M.; et al. Middle East Respiratory Syndrome Coronavirus Neutralising Serum Antibodies in Dromedary Camels: A Comparative Serological Study. Lancet Infect. Dis. 2013, 13, 859–866. [Google Scholar] [CrossRef]
- Hallmaier-Wacker, L.K.; Munster, V.J.; Knauf, S. Disease Reservoirs: From Conceptual Frameworks to Applicable Criteria. Emerg. Microbes Infect. 2017, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.J.; Seifert, S.N.; Carlson, C.J. Beyond Infection: Integrating Competence into Reservoir Host Prediction. Trends Ecol. Evol. 2020, 35, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and Coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, Evolution and Classification of Bat Coronaviruses in the Aftermath of SARS. Antiviral Res. 2014, 101, 45–56. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Mohd, H.A.; Al-Tawfiq, J.A.; Memish, Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Origin and Animal Reservoir. Virol. J. 2016, 13, 87. [Google Scholar] [CrossRef]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of Exposure to SARS-CoV-2 in Cats and Dogs from Households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Wood, J.L.N.; Leach, M.; Waldman, L.; MacGregor, H.; Fooks, A.R.; Jones, K.E.; Restif, O.; Dechmann, D.; Hayman, D.T.S.; Baker, K.S.; et al. A Framework for the Study of Zoonotic Disease Emergence and Its Drivers: Spillover of Bat Pathogens as a Case Study. Philos. Trans. R. Soc. B-Biol. Sci. 2012, 367, 2881–2892. [Google Scholar] [CrossRef]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A Molecular Phylogeny for Bats Illuminates Biogeography and the Fossil Record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef]
- Leroy, E.M.; Kumulungui, B.; Pourrut, X.; Rouquet, P.; Hassanin, A.; Yaba, P.; Delicat, A.; Paweska, J.T.; Gonzalez, J.P.; Swanepoel, R. Fruit Bats as Reservoirs of Ebola Virus. Nature 2005, 438, 575–576. [Google Scholar] [CrossRef]
- Pourrut, X.; Souris, M.; Towner, J.S.; Rollin, P.E.; Nichol, S.T.; Gonzalez, J.-P.; Leroy, E. Large Serological Survey Showing Cocirculation of Ebola and Marburg Viruses in Gabonese Bat Populations, and a High Seroprevalence of Both Viruses in Rousettus Aegyptiacus. BMC Infect. Dis. 2009, 9, 159. [Google Scholar] [CrossRef]
- Hayman, D.T.S.; Emmerich, P.; Yu, M.; Wang, L.-F.; Suu-Ire, R.; Fooks, A.R.; Cunningham, A.A.; Wood, J.L.N. Long-Term Survival of an Urban Fruit Bat Seropositive for Ebola and Lagos Bat Viruses. PLoS ONE 2010, 5, e11978. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T.S.; Yu, M.; Crameri, G.; Wang, L.-F.; Suu-Ire, R.; Wood, J.L.N.; Cunningham, A.A. Ebola Virus Antibodies in Fruit Bats, Ghana, West Africa. Emerg. Infect. Dis. 2012, 18, 1207–1209. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhang, Y.; Li, J.; Zhang, Y.; Wang, L.-F.; Shi, Z. Serological Evidence of Ebolavirus Infection in Bats, China. Virol. J. 2012, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Islam, A.; Yu, M.; Anthony, S.J.; Epstein, J.H.; Khan, S.A.; Khan, S.U.; Crameri, G.; Wang, L.-F.; Lipkin, W.I.; et al. Ebola Virus Antibodies in Fruit Bats, Bangladesh. Emerg. Infect. Dis. 2013, 19, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Miyamoto, H.; Nakayama, E.; Yoshida, R.; Nakamura, I.; Sawa, H.; Ishii, A.; Thomas, Y.; Nakagawa, E.; Matsuno, K.; et al. Seroepidemiological Prevalence of Multiple Species of Filoviruses in Fruit Bats (Eidolon helvum) Migrating in Africa. J. Infect. Dis. 2015, 212, S101–S108. [Google Scholar] [CrossRef]
- De Nys, H.M.; Kingebeni, P.M.; Keita, A.K.; Butel, C.; Thaurignac, G.; Villabona-Arenas, C.-J.; Lemarcis, T.; Geraerts, M.; Vidal, N.; Esteban, A.; et al. Survey of Ebola Viruses in Frugivorous and Insectivorous Bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015–2017. Emerg. Infect. Dis. 2018, 24, 2228–2240. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Bourgarel, M.; Cappelle, J.; Liégeois, F.; De Nys, H.M.; Roger, F. Ebola Virus Maintenance: If Not (Only) Bats, What Else? Viruses 2018, 10, 549. [Google Scholar] [CrossRef]
- Odening, K. Conception and terminology of hosts in parasitology. In Advances in Parasitology; Academic Press: London, UK, 1976; Volume 14, pp. 1–93. ISBN 978-0-12-031714-1. [Google Scholar]
- Parker, G.A.; Ball, M.A.; Chubb, J.C. Why Do Larval Helminths Avoid the Gut of Intermediate Hosts? J. Theor. Biol. 2009, 260, 460–473. [Google Scholar] [CrossRef]
- Plowright, R.K.; Eby, P.; Hudson, P.J.; Smith, I.L.; Westcott, D.; Bryden, W.L.; Middleton, D.; Reid, P.A.; McFarlane, R.A.; Martin, G.; et al. Ecological Dynamics of Emerging Bat Virus Spillover. Proc. R. Soc. B-Biol. Sci. 2015, 282, 20142124. [Google Scholar] [CrossRef] [PubMed]
- Goater, T.M.; Goater, C.P.; Esch, G.W. Parasitism: The Diversity and Ecology of Animal Parasites; Cambridge University Press: Cambridge, UK, 2014; ISBN 978-0-521-19028-2. [Google Scholar]
- Stigge, H.A.; Bolek, M.G. The Alteration of Life History Traits and Increased Success of Halipegus Eccentricus through the Use of a Paratenic Host: A Comparative Study. J. Parasitol. 2015, 101, 658–665. [Google Scholar] [CrossRef]
- Chua, K.B.; Lek Koh, C.; Hooi, P.S.; Wee, K.F.; Khong, J.H.; Chua, B.H.; Chan, Y.P.; Lim, M.E.; Lam, S.K. Isolation of Nipah Virus from Malaysian Island Flying-Foxes. Microbes Infect. 2002, 4, 145–151. [Google Scholar] [CrossRef]
- Reynes, J.-M.; Counor, D.; Ong, S.; Faure, C.; Seng, V.; Molia, S.; Walston, J.; Georges-Courbot, M.C.; Deubel, V.; Sarthou, J.-L. Nipah Virus in Lyle’s Flying Foxes, Cambodia. Emerg. Infect. Dis. 2005, 11, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.A.; Hassan, S.S.; Olival, K.J.; Mohamed, M.; Chang, L.-Y.; Hassan, L.; Saad, N.M.; Shohaimi, S.A.; Mamat, Z.C.; Naim, M.S.; et al. Characterization of Nipah Virus from Naturally Infected Pteropus Vampyrus Bats, Malaysia. Emerg. Infect. Dis. 2010, 16, 1990–1993. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Bellini, W.J.; Rota, P.A.; Harcourt, B.H.; Tamin, A.; Lam, S.K.; Ksiazek, T.G.; Rollin, P.E.; Zaki, S.R.; Shieh, W.-J.; et al. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science 2000, 288, 1432–1435. [Google Scholar] [CrossRef] [PubMed]
- Pulliam, J.R.C.; Epstein, J.H.; Dushoff, J.; Rahman, S.A.; Bunning, M.; Jamaluddin, A.A.; Hyatt, A.D.; Field, H.E.; Dobson, A.P.; Daszak, P. Agricultural Intensification, Priming for Persistence and the Emergence of Nipah Virus: A Lethal Bat-Borne Zoonosis. J. R. Soc. Interface 2011, 9, 89–101. [Google Scholar] [CrossRef]
- Luby, S.P.; Rahman, M.; Hossain, M.J.; Blum, L.S.; Husain, M.M.; Gurley, E.; Khan, R.; Ahmed, B.-N.; Rahman, S.; Nahar, N.; et al. Foodborne Transmission of Nipah Virus, Bangladesh. Emerg. Infect. Dis. 2006, 12, 1888–1894. [Google Scholar] [CrossRef]
- Rahman, M.A.; Hossain, M.J.; Sultana, S.; Homaira, N.; Khan, S.U.; Rahman, M.; Gurley, E.S.; Rollin, P.E.; Lo, M.K.; Comer, J.A.; et al. Date Palm Sap Linked to Nipah Virus Outbreak in Bangladesh, 2008. Vector-Borne Zoonotic Dis. 2011, 12, 65–72. [Google Scholar] [CrossRef]
- Reusken, C.B.; Ababneh, M.; Raj, V.S.; Meyer, B.; Eljarah, A.; Abutarbush, S.; Godeke, G.J.; Bestebroer, T.M.; Zutt, I.; Müller, M.A.; et al. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Serology in Major Livestock Species in an Affected Region in Jordan, June to September 2013. Eurosurveillance 2013, 18, 20662. [Google Scholar] [CrossRef]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; AlHakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F. Rooting the Phylogenetic Tree of Middle East Respiratory Syndrome Coronavirus by Characterization of a Conspecific Virus from an African Bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.M.; Said, M.Y.; Gluecks, I.; Lattwein, E.; Bosch, B.-J.; Drexler, J.F.; et al. Antibodies against MERS Coronavirus in Dromedary Camels, Kenya, 1992–2013. Emerg. Infect. Dis. 2014, 20, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Saqib, M.; Sieberg, A.; Hussain, M.H.; Mansoor, M.K.; Zohaib, A.; Lattwein, E.; Müller, M.A.; Drosten, C.; Corman, V.M. Serologic Evidence for MERS-CoV Infection in Dromedary Camels, Punjab, Pakistan, 2012–2015. Emerg. Infect. Dis. 2017, 23, 550–551. [Google Scholar] [CrossRef]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and Emergent Coronaviruses: Viral Determinants of Interspecies Transmission. Curr. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef]
- Epstein, J.H.; Field, H.E.; Luby, S.; Pulliam, J.R.C.; Daszak, P. Nipah Virus: Impact, Origins, and Causes of Emergence. Curr. Infect. Dis. Rep. 2006, 8, 59–65. [Google Scholar] [CrossRef]
- Caron, A.; Cappelle, J.; Cumming, G.S.; de Garine-Wichatitsky, M.; Gaidet, N. Bridge Hosts, a Missing Link for Disease Ecology in Multi-Host Systems. Vet. Res. 2015, 46, 83. [Google Scholar] [CrossRef]
- Wilson, A.J.; Morgan, E.R.; Booth, M.; Norman, R.; Perkins, S.E.; Hauffe, H.C.; Mideo, N.; Antonovics, J.; McCallum, H.; Fenton, A. What Is a Vector? Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160085. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.-J.J. Biological Transmission of Arboviruses: Reexamination of and New Insights into Components, Mechanisms, and Unique Traits as Well as Their Evolutionary Trends. Clin. Microbiol. Rev. 2005, 18, 608–637. [Google Scholar] [CrossRef]
- Levine, O.S.; Levine, M.M. Houseflies (Musca domestica) as Mechanical Vectors of Shigellosis. Rev. Infect. Dis. 1991, 13, 688–696. [Google Scholar] [CrossRef]
- CDC Division of Vector-Borne Diseases (DVBD). Available online: https://www.cdc.gov/ncezid/dvbd/index.html (accessed on 20 February 2021).
- ECDC Vector-Borne Diseases. Available online: https://www.ecdc.europa.eu/en/climate-change/climate-change-europe/vector-borne-diseases (accessed on 20 February 2021).
- Lafferty, K.D.; Kuris, A.M. Trophic Strategies, Animal Diversity and Body Size. Trends Ecol. Evol. 2002, 17, 507–513. [Google Scholar] [CrossRef]
- McCaughey, C.; Hart, C.A. Hantaviruses. J. Med. Microbiol. 2000, 49, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Müller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Primer 2017, 3, 17091. [Google Scholar] [CrossRef] [PubMed]
- WHO Vector-Borne Diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases (accessed on 20 February 2021).
- Vashi, N.A.; Reddy, P.; Wayne, D.B.; Sabin, B. Bat-Associated Leptospirosis. J. Gen. Intern. Med. 2010, 25, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Power, A.G.; Mitchell, C.E. Pathogen Spillover in Disease Epidemics. Am. Nat. 2004, 164, S79–S89. [Google Scholar] [CrossRef] [PubMed]
- Han, B.A.; Kramer, A.M.; Drake, J.M. Global Patterns of Zoonotic Disease in Mammals. Trends Parasitol. 2016, 32, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Parrish, C.R.; McCallum, H.; Hudson, P.J.; Ko, A.I.; Graham, A.L.; Lloyd-Smith, J.O. Pathways to Zoonotic Spillover. Nat. Rev. Microbiol. 2017, 15, 502–510. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Expert Consultation on Rabies: Third Report; World Health Organization: Geneva, Switzerland, 2018; ISBN 978-92-4-121021-8. [Google Scholar]
- Li, H.; Mendelsohn, E.; Zong, C.; Zhang, W.; Hagan, E.; Wang, N.; Li, S.; Yan, H.; Huang, H.; Zhu, G.; et al. Human-Animal Interactions and Bat Coronavirus Spillover Potential among Rural Residents in Southern China. Biosaf. Health 2019, 1, 84–90. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Johara, M.Y.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah Virus Infection in Bats (Order Chiroptera) in Peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar]
- Nuki, Paul Spillover: The Origins of Covid-19 and Why the next Pandemic May Already Have Started. The Telegraph, 2020. Available online: https://www.telegraph.co.uk/global-health/science-and-disease/spillover-origins-covid-19-next-pandemic-may-already-have-started/ (accessed on 20 February 2021).
- Becker, D.J.; Washburne, A.D.; Faust, C.L.; Pulliam, J.R.C.; Mordecai, E.A.; Lloyd-Smith, J.O.; Plowright, R.K. Dynamic and Integrative Approaches to Understanding Pathogen Spillover. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190014. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.G.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; et al. Genomic Surveillance Elucidates Ebola Virus Origin and Transmission during the 2014 Outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.W.; Paterson, R.A.; Townsend, C.R.; Poulin, R.; Tompkins, D.M. Parasite Spillback: A Neglected Concept in Invasion Ecology? Ecology 2009, 90, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Peña, A.; Ostfeld, R.; Peterson, A.; Poulin, R.; de la Fuente, J. Effects of Environmental Change on Zoonotic Disease Risk: An Ecological Primer. Trends Parasitol. 2014, 30. [Google Scholar] [CrossRef] [PubMed]
- Dobson, A.; Meagher, M. The Population Dynamics of Brucellosis in the Yellowstone National Park. Ecology 1996, 77, 1026–1036. [Google Scholar] [CrossRef]
- Daszak, P.; Cunningham, A.A.A.; Hyatt, A.D.D. Anthropogenic Environmental Change and the Emergence of Infectious Diseases in Wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef] [PubMed]
- VerCauteren, K.C.; Lavelle, M.J.; Campa, H.I. Persistent Spillback of Bovine Tuberculosis From White-Tailed Deer to Cattle in Michigan, USA: Status, Strategies, and Needs. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Donnelly, C.A.; Nouvellet, P. The Contribution of Badgers to Confirmed Tuberculosis in Cattle in High-Incidence Areas in England. PLOS Curr. Outbreaks 2013. [Google Scholar] [CrossRef]
- Patterson, B.D.; Ramírez-Chaves, H.E.; Vilela, J.F.; Soares, A.E.R.; Grewe, F. and On the Nomenclature of the American Clade of Weasels (Carnivora: Mustelidae). J. Anim. Divers. 2021, 3. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on Mink Farms between Humans and Mink and Back to Humans. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Fagre, A.; Cohen, L.E.; Eskew, E.A.; Farrell, M.; Glennon, E.; Joseph, M.B.; Frank, H.K.; Ryan, S.J.; Carlson, C.J.; Albery, G. Spillback in the Anthropocene: The Risk of Human-to-Wildlife Pathogen Transmission for Conservation and Public Health. EcoEvoRxiv 2021. [Google Scholar] [CrossRef]
- Olival, K.J.; Cryan, P.M.; Amman, B.R.; Baric, R.S.; Blehert, D.S.; Brook, C.E.; Calisher, C.H.; Castle, K.T.; Coleman, J.T.H.; Daszak, P.; et al. Possibility for Reverse Zoonotic Transmission of SARS-CoV-2 to Free-Ranging Wildlife: A Case Study of Bats. PLOS Pathog. 2020, 16, e1008758. [Google Scholar] [CrossRef]
- Fagre, A.; Lewis, J.; Eckley, M.; Zhan, S.; Rocha, S.M.; Sexton, N.R.; Burke, B.; Geiss, B.; Peersen, O.; Kading, R.; et al. SARS-CoV-2 Infection, Neuropathogenesis and Transmission among Deer Mice: Implications for Reverse Zoonosis to New World Rodents. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cunningham, A.; Daszak, P.; Rodríguez, J. Pathogen Pollution: Defining a Parasitological Threat to Biodiversity Conservation. J Parasitol. 2003, 89, S78–S83. [Google Scholar]
- Grubaugh, N.D.; Petrone, M.E.; Holmes, E.C. We Shouldn’t Worry When a Virus Mutates during Disease Outbreaks. Nat. Microbiol. 2020, 5, 529–530. [Google Scholar] [CrossRef]
- Lauring, A.S.; Frydman, J.; Andino, R. The Role of Mutational Robustness in RNA Virus Evolution. Nat. Rev. Microbiol. 2013, 11, 327–336. [Google Scholar] [CrossRef]
- Gómez, C.E.; Perdiguero, B.; Esteban, M. Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs against SARS-CoV-2/COVID-19. Vaccines 2021, 9, 243. [Google Scholar] [CrossRef]
- Public Health England. Investigation of Novel SARS-COV-2 Variant: Variant of Concern 202012/01; Public Health England: London, UK, 2020. Available online: https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 (accessed on 20 February 2021).
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; da Candido, D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and Epidemiology of a Novel SARS-CoV-2 Lineage in Manaus, Brazil. medRxiv 2021. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Lineage with Multiple Spike Mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Darwin, C. The Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. Nature 1872, 5, 318–319. [Google Scholar]
- Swaroop, S. Index of Endemicity. Bull. World Health Organ. 1957, 16, 1083–1101. [Google Scholar]
- Merrill, R.M.; Timmreck, T.C. Introduction to Epidemiology; Jones and Bartlett Publishers: Sudbury, MA, USA, 2006; ISBN 978-0-7637-3582-1. [Google Scholar]
- CDC Principles of Epidemiology: Lesson 1—Section 11. Available online: https://www.cdc.gov/csels/dsepd/ss1978/lesson1/section11.html (accessed on 23 December 2020).
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D.T. Out of Africa: A Molecular Perspective on the Introduction of Yellow Fever Virus into the Americas. PLOS Pathog. 2007, 3, e75. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.T.; Wearing, H.J.; Jr, R.C.R.; Morrison, A.C.; Astete, H.; Vilcarromero, S.; Alvarez, C.; Ramal-Asayag, C.; Sihuincha, M.; Rocha, C.; et al. Long-Term and Seasonal Dynamics of Dengue in Iquitos, Peru. PLoS Negl. Trop. Dis. 2014, 8, e3003. [Google Scholar] [CrossRef] [PubMed]
- Wobeser, G.A. Disease and Epizootiology—Basic Principles. In Investigation and Management of Disease in Wild Animals; Wobeser, G.A., Ed.; Springer: Boston, MA, USA, 1994; pp. 3–12. ISBN 978-1-4757-5609-8. [Google Scholar]
- Ren, W.; Qu, X.; Li, W.; Han, Z.; Yu, M.; Zhou, P.; Zhang, S.-Y.; Wang, L.-F.; Deng, H.; Shi, Z. Difference in Receptor Usage between Severe Acute Respiratory Syndrome (SARS) Coronavirus and SARS-Like Coronavirus of Bat Origin. J. Virol. 2008, 82, 1899–1907. [Google Scholar] [CrossRef]
- Yaiw, K.C.; Bingham, J.; Crameri, G.; Mungall, B.; Hyatt, A.; Yu, M.; Eaton, B.; Shamala, D.; Wang, L.-F.; Wong, K.T. Tioman Virus, a Paramyxovirus of Bat Origin, Causes Mild Disease in Pigs and Has a Predilection for Lymphoid Tissues. J. Virol. 2008, 82, 565–568. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El Zowalaty, M.E.; Järhult, J.D. From SARS to COVID-19: A Previously Unknown SARS- Related Coronavirus (SARS-CoV-2) of Pandemic Potential Infecting Humans—Call for a One Health Approach. One Health 2020, 9, 100124. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Bininda-Emonds, O.R.P.; Gittleman, J.L. Bats, Clocks, and Rocks: Diversification Patterns in Chiroptera. Evolution 2005, 59, 2243–2255. [Google Scholar] [CrossRef]
- Campbell, C.J.; Nelson, D.M.; Ogawa, N.O.; Chikaraishi, Y.; Ohkouchi, N. Trophic Position and Dietary Breadth of Bats Revealed by Nitrogen Isotopic Composition of Amino Acids. Sci. Rep. 2017, 7, 15932. [Google Scholar] [CrossRef]
- Findley, J.S.; Wilson, D.E. Ecological Significance of Chiropteran Morphology. In Ecology of Bats; Kunz, T.H., Ed.; Springer: Boston, MA, USA, 1982; pp. 243–260. ISBN 978-1-4613-3423-1. [Google Scholar]
- Patterson, B.; Willig, M.; Stevens, R. Trophic strategies, niche partitioning, and patterns of ecological organization. In Bat Ecology; Kunz, T.H., Fenton, B., Eds.; University of Chicago Press: Chicago, IL, USA, 2003; pp. 536–579. [Google Scholar]
- Fenton, M.B. Science and the Conservation of Bats. J. Mammal. 1997, 78, 1–14. [Google Scholar] [CrossRef]
- Monadjem, A.; Reside, A.; Cornut, J.; Perrin, M.R. Roost Selection and Home Range of an African Insectivorous Bat Nycteris Thebaica (Chiroptera, Nycteridae). Mammalia 2009, 73, 353–359. [Google Scholar] [CrossRef]
- Fleming, T.H. Bat Migration. Encycl. Anim. Behav. 2019, 605. [Google Scholar] [CrossRef]
- Wilkinson, G.S. Social and vocal complexity in bats. In Animal Social Complexity: Intelligence, Culture, and Individualized Societies; Harvard University Press: Cambridge, UK, 2003; pp. 322–341. [Google Scholar]
- Kunz, T.H. Roosting Ecology of Bats. In Ecology of Bats; Kunz, T.H., Ed.; Springer: Boston, MA, USA, 1982; pp. 1–55. ISBN 978-1-4613-3421-7. [Google Scholar]
- Barclay, R.M.R.; Ulmer, J.; MacKenzie, C.J.A.; Thompson, M.S.; Olson, L.; McCool, J.; Cropley, E.; Poll, G. Variation in the Reproductive Rate of Bats. Can. J. Zool. 2004, 82, 688–693. [Google Scholar] [CrossRef]
- Brunet-Rossinni, A.K.; Austad, S.N. Ageing Studies on Bats: A Review. Biogerontology 2004, 5, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Brook, C.E.; Dobson, A.P. Bats as ‘Special’ Reservoirs for Emerging Zoonotic Pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Kalko, E.K.V. Where Forest Meets Urbanization: Foraging Plasticity of Aerial Insectivorous Bats in an Anthropogenically Altered Environment. J. Mammal. 2010, 91, 144–153. [Google Scholar] [CrossRef]
- Threlfall, C.G.; Law, B.; Banks, P.B. Sensitivity of Insectivorous Bats to Urbanization: Implications for Suburban Conservation Planning. Biol. Conserv. 2012, 146, 41–52. [Google Scholar] [CrossRef]
- Russo, D.; Ancillotto, L. Sensitivity of Bats to Urbanization: A Review. Mamm. Biol. 2015, 80, 205–212. [Google Scholar] [CrossRef]
- Mickleburgh, S.; Waylen, K.; Racey, P. Bats as Bushmeat: A Global Review. Oryx 2009, 43, 217–234. [Google Scholar] [CrossRef]
- Mildenstein, T.; Tanshi, I.; Racey, P.A. Exploitation of Bats for Bushmeat and Medicine. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Christian, V., Tigga, K., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 325–375. [Google Scholar]
- Willoughby, A.R.; Phelps, K.L.; PREDICT Consortium; Olival, K.J. A Comparative Analysis of Viral Richness and Viral Sharing in Cave-Roosting Bats. Diversity 2017, 9, 35. [Google Scholar] [CrossRef]
- Ridley, M. The Bats behind the Pandemic. Wall Str. J. Available online: https://www.wsj.com/articles/the-bats-behind-the-pandemic-11586440959 (accessed on 13 June 2021).
- Gilbert, A.T.; Fooks, A.R.; Hayman, D.T.S.; Horton, D.L.; Müller, T.; Plowright, R.; Peel, A.J.; Bowen, R.; Wood, J.L.N.; Mills, J.; et al. Deciphering Serology to Understand the Ecology of Infectious Diseases in Wildlife. EcoHealth 2013, 10, 298–313. [Google Scholar] [CrossRef]
- Becker, D.J.; Broos, A.; Bergner, L.M.; Meza, D.K.; Simmons, N.B.; Fenton, M.B.; Altizer, S.; Streicker, D.G. Temporal Patterns of Vampire Bat Rabies and Host Connectivity in Belize. Transbound. Emerg. Dis. 2021, 68, 870–879. [Google Scholar] [CrossRef]
- Amman, B.R.; Carroll, S.A.; Reed, Z.D.; Sealy, T.K.; Balinandi, S.; Swanepoel, R.; Kemp, A.; Erickson, B.R.; Comer, J.A.; Campbell, S.; et al. Seasonal Pulses of Marburg Virus Circulation in Juvenile Rousettus aegyptiacus Bats Coincide with Periods of Increased Risk of Human Infection. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Peel, A.J.; McKinley, T.J.; Baker, K.S.; Barr, J.A.; Crameri, G.; Hayman, D.T.S.; Feng, Y.-R.; Broder, C.C.; Wang, L.-F.; Cunningham, A.A.; et al. Use of Cross-Reactive Serological Assays for Detecting Novel Pathogens in Wildlife: Assessing an Appropriate Cutoff for Henipavirus Assays in African Bats. J. Virol. Methods 2013, 193, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Dovih, P.; Laing, E.D.; Chen, Y.; Low, D.H.W.; Ansil, B.R.; Yang, X.; Shi, Z.; Broder, C.C.; Smith, G.J.D.; Linster, M.; et al. Filovirus-Reactive Antibodies in Humans and Bats in Northeast India Imply Zoonotic Spillover. PLoS Negl. Trop. Dis. 2019, 13, e0007733. [Google Scholar] [CrossRef]
- Calisher, C.H.; Karabatsos, N.; Dalrymple, J.M.; Shope, R.E.; Porterfield, J.S.; Westaway, E.G.; Brandt, W.E. Antigenic Relationships between Flaviviruses as Determined by Cross-Neutralization Tests with Polyclonal Antisera. J. Gen. Virol. 1989, 70 Pt 1, 37–43. [Google Scholar] [CrossRef]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra Virus from Pteropid Bats: A Natural Reservoir of Hendra Virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Niezgoda, M.; Franka, R.; Agwanda, B.; Markotter, W.; Beagley, J.C.; Urazova, O.Y.; Breiman, R.F.; Rupprecht, C.E. Possible Emergence of West Caucasian Bat Virus in Africa. Emerg. Infect. Dis. 2008, 14, 1887–1889. [Google Scholar] [CrossRef]
- Epstein, J.H.; Baker, M.L.; Zambrana-Torrelio, C.; Middleton, D.; Barr, J.A.; DuBovi, E.; Boyd, V.; Pope, B.; Todd, S.; Crameri, G.; et al. Duration of Maternal Antibodies against Canine Distemper Virus and Hendra Virus in Pteropid Bats. PLoS ONE 2013, 8, e67584. [Google Scholar] [CrossRef]
- Baker, K.S.; Suu-Ire, R.; Barr, J.; Hayman, D.T.S.; Broder, C.C.; Horton, D.L.; Durrant, C.; Murcia, P.R.; Cunningham, A.A.; Wood, J.L.N. Viral Antibody Dynamics in a Chiropteran Host. J. Anim. Ecol. 2014, 83, 415–428. [Google Scholar] [CrossRef]
- Maruyama, J.; Miyamoto, H.; Kajihara, M.; Ogawa, H.; Maeda, K.; Sakoda, Y.; Yoshida, R.; Takada, A. Characterization of the Envelope Glycoprotein of a Novel Filovirus, Lloviu Virus. J. Virol. 2014, 88, 99–109. [Google Scholar] [CrossRef]
- Leopardi, S.; Priori, P.; Zecchin, B.; Poglayen, G.; Trevisiol, K.; Lelli, D.; Zoppi, S.; Scicluna, M.T.; D’Avino, N.; Schiavon, E.; et al. Active and Passive Surveillance for Bat Lyssaviruses in Italy Revealed Serological Evidence for Their Circulation in Three Bat Species. Epidemiol. Infect. 2019, 147. [Google Scholar] [CrossRef]
- Brook, C.E.; Ranaivoson, H.C.; Broder, C.C.; Cunningham, A.A.; Héraud, J.-M.; Peel, A.J.; Gibson, L.; Wood, J.L.N.; Metcalf, C.J.; Dobson, A.P. Disentangling Serology to Elucidate Henipa- and Filovirus Transmission in Madagascar Fruit Bats. J. Anim. Ecol. 2019, 88, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, A.; Smreczak, M.; Freuling, C.M.; Müller, T.; Trębas, P.; Rola, J. Serological Survey of Lyssaviruses in Polish Bats in the Frame of Passive Rabies Surveillance Using an Enzyme-Linked Immunosorbent Assay. Viruses 2020, 12, 271. [Google Scholar] [CrossRef] [PubMed]
- Peel, A.J.; Baker, K.S.; Hayman, D.T.S.; Broder, C.C.; Cunningham, A.A.; Fooks, A.R.; Garnier, R.; Wood, J.L.N.; Restif, O. Support for Viral Persistence in Bats from Age-Specific Serology and Models of Maternal Immunity. Sci. Rep. 2018, 8, 3859. [Google Scholar] [CrossRef] [PubMed]
- Glennon, E.E.; Becker, D.J.; Peel, A.J.; Garnier, R.; Suu-Ire, R.D.; Gibson, L.; Hayman, D.T.S.; Wood, J.L.N.; Cunningham, A.A.; Plowright, R.K.; et al. What Is Stirring in the Reservoir? Modelling Mechanisms of Henipavirus Circulation in Fruit Bat Hosts. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190021. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, T.J.; Bowen, R.A.; Stanley, T.R.; Shankar, V.; Rupprecht, C.E. Variability in Seroprevalence of Rabies Virus Neutralizing Antibodies and Associated Factors in a Colorado Population of Big Brown Bats (Eptesicus fuscus). PLoS ONE 2014, 9, e86261. [Google Scholar] [CrossRef]
- Ogunkoya, A.B.; Beran, G.W.; Umoh, J.U.; Gomwalk, N.E.; Abdulkadir, I.A. Serological Evidence of Infection of Dogs and Man in Nigeria by Lyssaviruses (Family Rhabdoviridae). Trans. R. Soc. Trop. Med. Hyg. 1990, 84, 842–845. [Google Scholar] [CrossRef]
- Benavides, J.A.; Velasco-Villa, A.; Godino, L.C.; Satheshkumar, P.S.; Nino, R.; Rojas-Paniagua, E.; Shiva, C.; Falcon, N.; Streicker, D.G. Abortive Vampire Bat Rabies Infections in Peruvian Peridomestic Livestock. PLoS Negl. Trop. Dis. 2020, 14, e0008194. [Google Scholar] [CrossRef]
- Thiermann, A.B. Bovine Leptospirosis: Bacteriologic versus Serologic Diagnosis of Cows at Slaughter. Am. J. Vet. Res. 1983, 44, 2244–2245. [Google Scholar] [PubMed]
- Otaka, D.; Martins, G.; Hamond, C.; Penna, B.; Medeiros, M.; Lilenbaum, W. Serology and PCR for Bovine Leptospirosis: Herd and Individual Approaches. Vet. Rec. 2012, 170, 338. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Calisher, C.H.; Dietzgen, R.G.; Kondo, H.; Kurath, G.; Longdon, B.; Stone, D.M.; Tesh, R.B.; Tordo, N.; et al. ICTV Virus Taxonomy Profile: Rhabdoviridae. J. Gen. Virol. 2018, 99, 447–448. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; Amengual, B.; Abellán, C.; Bourhy, H. European Bat Lyssavirus Infection in Spanish Bat Populations. Emerg. Infect. Dis. 2002, 8, 413–420. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; López-Roig, M.; Seguí, M.; Sánchez, L.P.; Nadal, J.; Borrás, M.; Lavenir, R.; Bourhy, H. Ecological Factors Associated with European Bat Lyssavirus Seroprevalence in Spanish Bats. PLoS ONE 2013, 8, e64467. [Google Scholar] [CrossRef]
- Storm, N.; Jansen Van Vuren, P.; Markotter, W.; Paweska, J.T. Antibody Responses to Marburg Virus in Egyptian Rousette Bats and Their Role in Protection against Infection. Viruses 2018, 10, 73. [Google Scholar] [CrossRef]
- Peel, A.J.; Baker, K.S.; Crameri, G.; Barr, J.A.; Hayman, D.T.S.; Wright, E.; Broder, C.C.; Fernández-Loras, A.; Fooks, A.R.; Wang, L.-F.; et al. Henipavirus Neutralising Antibodies in an Isolated Island Population of African Fruit Bats. PLoS ONE 2012, 7, e30346. [Google Scholar] [CrossRef] [PubMed]
- Kading, R.C.; Kityo, R.M.; Mossel, E.C.; Borland, E.M.; Nakayiki, T.; Nalikka, B.; Nyakarahuka, L.; Ledermann, J.P.; Panella, N.A.; Gilbert, A.T.; et al. Neutralizing Antibodies against Flaviviruses, Babanki Virus, and Rift Valley Fever Virus in Ugandan Bats. Infect. Ecol. Epidemiol. 2018, 8, 1439215. [Google Scholar] [CrossRef]
- Wang, L.-F.; Anderson, D.E. Viruses in Bats and Potential Spillover to Animals and Humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Wiens, J.J. The Role of Morphological Data in Phylogeny Reconstruction. Syst. Biol. 2004, 53, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Yang, Z.; Telford, M.J. Phylogenetic Tree Building in the Genomic Age. Nat. Rev. Genet. 2020, 21, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian Inference of Phylogeny and Its Impact on Evolutionary Biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Mavian, C.; Pond, S.K.; Marini, S.; Magalis, B.R.; Vandamme, A.-M.; Dellicour, S.; Scarpino, S.V.; Houldcroft, C.; Villabona-Arenas, J.; Paisie, T.K.; et al. Sampling Bias and Incorrect Rooting Make Phylogenetic Network Tracing of SARS-COV-2 Infections Unreliable. Proc. Natl. Acad. Sci. USA 2020, 117, 12522–12523. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- MacLean, O.A.; Lytras, S.; Weaver, S.; Singer, J.B.; Boni, M.F.; Lemey, P.; Pond, S.L.K.; Robertson, D.L. Natural Selection in the Evolution of SARS-CoV-2 in Bats Created a Generalist Virus and Highly Capable Human Pathogen. PLoS Biol. 2021, 19, e3001115. [Google Scholar] [CrossRef]
- Koyama, T.; Platt, D.; Parida, L. Variant Analysis of SARS-CoV-2 Genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Yang, Z.; Rannala, B. Molecular Phylogenetics: Principles and Practice. Nat. Rev. Genet. 2012, 13, 303–314. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, T. Bayesian Selection of Misspecified Models Is Overconfident and May Cause Spurious Posterior Probabilities for Phylogenetic Trees. Proc. Natl. Acad. Sci. USA 2018, 115, 1854–1859. [Google Scholar] [CrossRef]
- Nascimento, F.F.; dos Reis, M.; Yang, Z. A Biologist’s Guide to Bayesian Phylogenetic Analysis. Nat. Ecol. Evol. 2017, 1, 1446–1454. [Google Scholar] [CrossRef]
- Wiley, E.O.; Lieberman, B.S. Phylogenetics: Theory and Practice of Phylogenetic Systematics, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2011. [Google Scholar]
- Klopfstein, S.; Massingham, T.; Goldman, N. More on the Best Evolutionary Rate for Phylogenetic Analysis. Syst. Biol. 2017, 66, 769–785. [Google Scholar] [CrossRef]
- Lai, M.M. RNA Recombination in Animal and Plant Viruses. Microbiol. Rev. 1992, 56, 61–79. [Google Scholar] [CrossRef]
- Ji, W.; Wang, W.; Zhao, X.; Zai, J.; Li, X. Cross-Species Transmission of the Newly Identified Coronavirus 2019-NCoV. J. Med. Virol. 2020, 92, 433–440. [Google Scholar] [CrossRef]
- Scheirer, W. A Pandemic of Bad Science. Bull. At. Sci. 2020, 76, 175–184. [Google Scholar] [CrossRef]
- Callaway, E.; Cyranoski, D. Why Snakes Probably Aren’t Spreading the New China Virus. Nature 2020. [Google Scholar] [CrossRef]
- Carlson, C.J.; Zipfel, C.M.; Garnier, R.; Bansal, S. Global Estimates of Mammalian Viral Diversity Accounting for Host Sharing. Nat. Ecol. Evol. 2019, 3, 1070–1075. [Google Scholar] [CrossRef]
- Anthony, S.J.; Epstein, J.H.; Murray, K.A.; Navarrete-Macias, I.; Zambrana-Torrelio, C.M.; Solovyov, A.; Ojeda-Flores, R.; Arrigo, N.C.; Islam, A.; Khan, S.A.; et al. A Strategy to Estimate Unknown Viral Diversity in Mammals. mBio 2013, 4. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Chia, W.N.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 Related Coronaviruses Circulating in Bats and Pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef] [PubMed]
- Sokolow, S.H.; Nova, N.; Pepin, K.M.; Peel, A.J.; Pulliam, J.R.C.; Manlove, K.; Cross, P.C.; Becker, D.J.; Plowright, R.K.; McCallum, H.; et al. Ecological Interventions to Prevent and Manage Zoonotic Pathogen Spillover. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180342. [Google Scholar] [CrossRef] [PubMed]
- Nugent, G. Maintenance, Spillover and Spillback Transmission of Bovine Tuberculosis in Multi-Host Wildlife Complexes: A New Zealand Case Study. Vet. Microbiol. 2011, 151, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Gurley, E.S.; Hossain, M.J.; Nahar, N.; Sharker, M.A.Y.; Luby, S.P. A Randomized Controlled Trial of Interventions to Impede Date Palm Sap Contamination by Bats to Prevent Nipah Virus Transmission in Bangladesh. PLoS ONE 2012, 7, e42689. [Google Scholar] [CrossRef]
- Rocha, R.; Aziz, S.A.; Brook, C.E.; Carvalho, W.D.; Cooper-Bohannon, R.; Frick, W.F.; Huang, J.C.-C.; Kingston, T.; López-Baucells, A.; Maas, B.; et al. Bat Conservation and Zoonotic Disease Risk: A Research Agenda to Prevent Misguided Persecution in the Aftermath of COVID-19. Anim. Conserv. 2020. [Google Scholar] [CrossRef]
- Zhao, H. COVID-19 Drives New Threat to Bats in China. Science 2020, 367, 1436. [Google Scholar] [CrossRef] [PubMed]
- Manlove, K.R.; Walker, J.G.; Craft, M.E.; Huyvaert, K.P.; Joseph, M.B.; Miller, R.S.; Nol, P.; Patyk, K.A.; O’Brien, D.; Walsh, D.P.; et al. “One Health” or Three? Publication Silos among the One Health Disciplines. PLoS Biol. 2016, 14, e1002448. [Google Scholar] [CrossRef] [PubMed]
- Guth, S.; Visher, E.; Boots, M.; Brook, C.E. Host Phylogenetic Distance Drives Trends in Virus Virulence and Transmissibility across the Animal–Human Interface. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190296. [Google Scholar] [CrossRef]
- Sumner, P.; Vivian-Griffiths, S.; Boivin, J.; Williams, A.; Venetis, C.A.; Davies, A.; Ogden, J.; Whelan, L.; Hughes, B.; Dalton, B.; et al. The Association between Exaggeration in Health Related Science News and Academic Press Releases: Retrospective Observational Study. BMJ 2014, 349, g7015. [Google Scholar] [CrossRef]
- Wihbey, J. The Challenges of Democratizing News and Information: Examining Data on Social Media, Viral Patterns and Digital Influence. SSRN Electron. J. 2014. [Google Scholar] [CrossRef]
- Tripathy, J.P.; Bhatnagar, A.; Shewade, H.D.; Kumar, A.M.V.; Zachariah, R.; Harries, A.D. Ten Tips to Improve the Visibility and Dissemination of Research for Policy Makers and Practitioners. Public Health Action 2017, 7, 10–14. [Google Scholar] [CrossRef]
- Origgi, G.; Branch-Smith, T.; Morisseau, T. Trust, Expertise and the Controversy over Chloroquine. Soc. Epistemol. J. Knowl. Cult. Policy 2021. Available online: https://cen.acs.org/policy/global-health/Will-public-trust-in-science-survive-the-pandemic/99/i3 (accessed on 20 February 2021).
- Gollust, S.E.; Fowler, E.F.; Niederdeppe, J. Television News Coverage of Public Health Issues and Implications for Public Health Policy and Practice. Annu. Rev. Public Health 2019, 40, 167–185. [Google Scholar] [CrossRef]
- Gallotti, R.; Valle, F.; Castaldo, N.; Sacco, P.; De Domenico, M. Assessing the Risks of ‘Infodemics’ in Response to COVID-19 Epidemics. Nat. Hum. Behav. 2020, 4, 1285–1293. [Google Scholar] [CrossRef] [PubMed]

| Miscommunication Type | Source of Confusion | By | Examples | Solution | Implemented by |
|---|---|---|---|---|---|
| 1 | Incorrect or overly broad use | Messenger | Pathogen Reservoir Intermediate host Spillback Bats | Confirm definitions in the literature | Messenger |
| 2 | Unstable usage or different usage between disciplines | Messenger and/or Receiver | Intermediate host Vector Endemic Bat origin | Provide definition in text Use more specific alternatives if available | Messenger |
| 3 | Incorrect inference about biological process | Receiver | Spillover Mutation Novel Bat origin | Avoid sensationalism Add precautionary language | Messenger and Receiver Messenger |
| 4 | Incorrect conclusion from evidence | Messenger and/or Receiver | Serology Phylogeny | Familiarize with different methods | Messenger and Receiver |
| Do not overinterpret findings | Messenger and Receiver | ||||
| Collaborate with experts | Messenger |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapiro, J.T.; Víquez-R, L.; Leopardi, S.; Vicente-Santos, A.; Mendenhall, I.H.; Frick, W.F.; Kading, R.C.; Medellín, R.A.; Racey, P.; Kingston, T. Setting the Terms for Zoonotic Diseases: Effective Communication for Research, Conservation, and Public Policy. Viruses 2021, 13, 1356. https://doi.org/10.3390/v13071356
Shapiro JT, Víquez-R L, Leopardi S, Vicente-Santos A, Mendenhall IH, Frick WF, Kading RC, Medellín RA, Racey P, Kingston T. Setting the Terms for Zoonotic Diseases: Effective Communication for Research, Conservation, and Public Policy. Viruses. 2021; 13(7):1356. https://doi.org/10.3390/v13071356
Chicago/Turabian StyleShapiro, Julie Teresa, Luis Víquez-R, Stefania Leopardi, Amanda Vicente-Santos, Ian H. Mendenhall, Winifred F. Frick, Rebekah C. Kading, Rodrigo A. Medellín, Paul Racey, and Tigga Kingston. 2021. "Setting the Terms for Zoonotic Diseases: Effective Communication for Research, Conservation, and Public Policy" Viruses 13, no. 7: 1356. https://doi.org/10.3390/v13071356
APA StyleShapiro, J. T., Víquez-R, L., Leopardi, S., Vicente-Santos, A., Mendenhall, I. H., Frick, W. F., Kading, R. C., Medellín, R. A., Racey, P., & Kingston, T. (2021). Setting the Terms for Zoonotic Diseases: Effective Communication for Research, Conservation, and Public Policy. Viruses, 13(7), 1356. https://doi.org/10.3390/v13071356







