How Do Flaviviruses Hijack Host Cell Functions by Phase Separation?
Abstract
:1. Introduction
2. Disordered Regions in Viral Proteins
2.1. Viral Proteins Are Rich in Disordered Regions
2.2. Advantages of High IDR Content in Viral Strategies
2.3. RNA Chaperone Activity of Viral IDRs
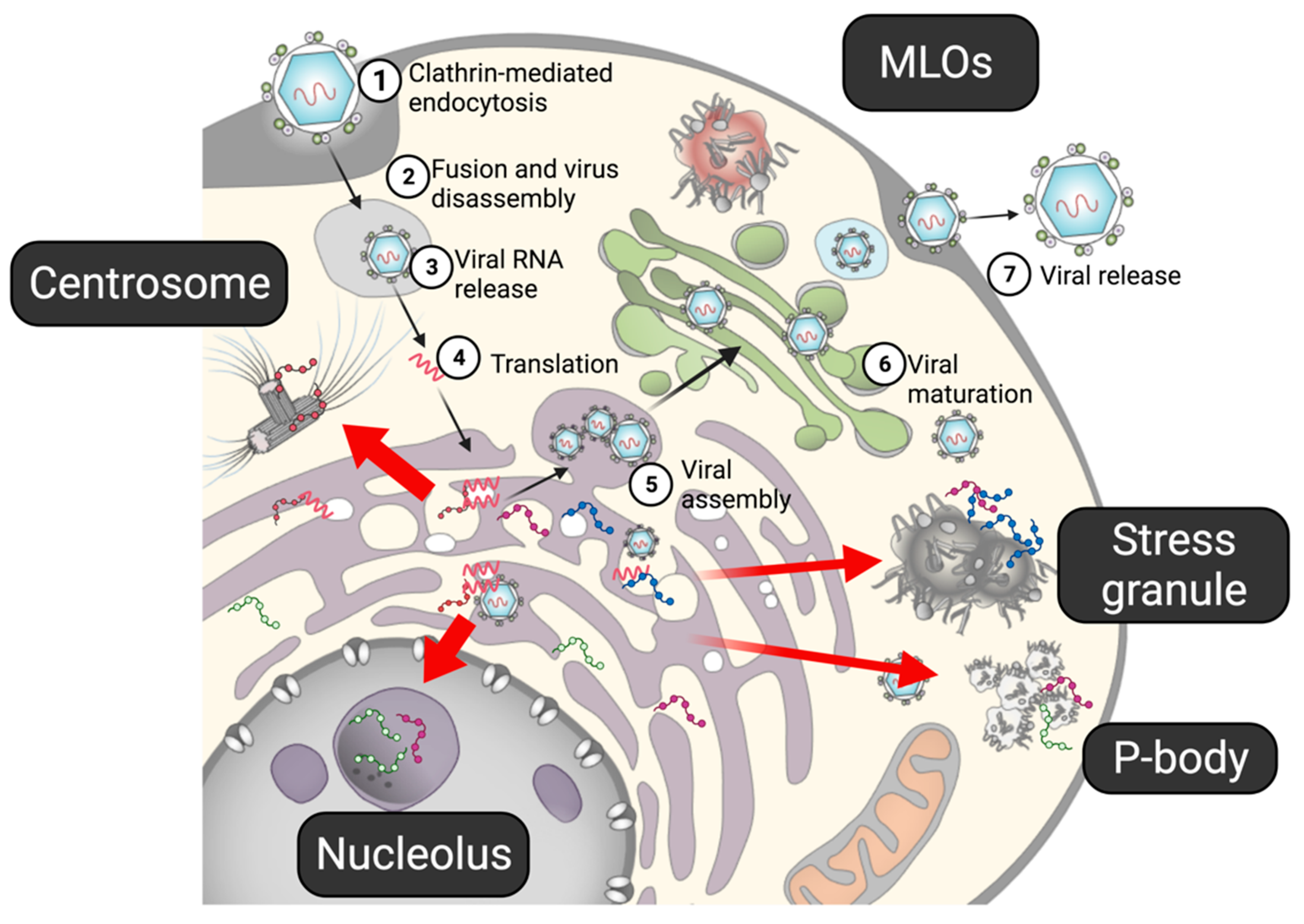
3. Impact of Intracellular MLOs on the Flaviviral Life Cycle
3.1. Flaviviral Proteins in Nucleolus
3.2. Assembly/Dissolution of SGs during Flaviviral Infection
3.3. Functional Interaction between Viral Proteins and SGs
4. Closing Remarks and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tokuriki, N.; Oldfield, C.J.; Uversky, V.N.; Berezovsky, I.N.; Tawfik, D.S. Do Viral Proteins Possess Unique Biophysical Features? Trends Biochem. Sci. 2009, 34, 53–59. [Google Scholar] [CrossRef]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Goh, G.K.-M.; Dunker, A.K.; Uversky, V.N. Viral Disorder or Disordered Viruses: Do Viral Proteins Possess Unique Features? Protein Pept. Lett. 2010, 17, 932–951. [Google Scholar] [CrossRef]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly Order in Protein Intrinsic Disorder Distribution: Disorder in 3500 Proteomes from Viruses and the Three Domains of Life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef]
- Tokunaga, M.; Miyamoto, Y.; Suzuki, T.; Otani, M.; Inuki, S.; Esaki, T.; Nagao, C.; Mizuguchi, K.; Ohno, H.; Yoneda, Y.; et al. Novel Anti-Flavivirus Drugs Targeting the Nucleolar Distribution of Core Protein. Virology 2020, 541, 41–51. [Google Scholar] [CrossRef]
- Fraser, J.E.; Rawlinson, S.M.; Heaton, S.M.; Jans, D.A. Dynamic Nucleolar Targeting of Dengue Virus Polymerase NS5 in Response to Extracellular PH. J. Virol. 2016, 90, 5797–5807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, Z.; Pan, T.; Wu, X.; Song, W.; Wang, S.; Xu, Y.; Rice, C.M.; MacDonald, M.R.; Yuan, Z. Hepatitis C Virus Co-Opts Ras-GTPase-Activating Protein-Binding Protein 1 for Its Genome Replication. J. Virol. 2011, 85, 6996–7004. [Google Scholar] [CrossRef] [Green Version]
- Hiscox, J.A.; Wurm, T.; Wilson, L.; Britton, P.; Cavanagh, D.; Brooks, G. The Coronavirus Infectious Bronchitis Virus Nucleoprotein Localizes to the Nucleolus. J. Virol. 2001, 75, 506–512. [Google Scholar] [CrossRef] [Green Version]
- You, J.-H.; Reed, M.L.; Hiscox, J.A. Trafficking Motifs in the SARS-Coronavirus Nucleocapsid Protein. Biochem. Biophys. Res. Commun. 2007, 358, 1015–1020. [Google Scholar] [CrossRef]
- Acar, D.D.; Stroobants, V.J.E.; Favoreel, H.; Saelens, X.; Nauwynck, H.J.Y. 2019 Identification of Peptide Domains Involved in the Subcellular Localization of the Feline Coronavirus 3b Protein. J. Gen. Virol. 2019, 100, 1417–1430. [Google Scholar] [CrossRef]
- Yuan, X.; Yao, Z.; Shan, Y.; Chen, B.; Yang, Z.; Wu, J.; Zhao, Z.; Chen, J.; Cong, Y. Nucleolar Localization of Non-Structural Protein 3b, a Protein Specifically Encoded by the Severe Acute Respiratory Syndrome Coronavirus. Virus Res. 2005, 114, 70–79. [Google Scholar] [CrossRef]
- Aminev, A.G.; Amineva, S.P.; Palmenberg, A.C. Encephalomyocarditis Viral Protein 2A Localizes to Nucleoli and Inhibits Cap-Dependent MRNA Translation. Virus Res. 2003, 95, 45–57. [Google Scholar] [CrossRef]
- Katoh, H.; Okamoto, T.; Fukuhara, T.; Kambara, H.; Morita, E.; Mori, Y.; Kamitani, W.; Matsuura, Y. Japanese Encephalitis Virus Core Protein Inhibits Stress Granule Formation through an Interaction with Caprin-1 and Facilitates Viral Propagation. J. Virol. 2013, 87, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Emara, M.M.; Brinton, M.A. Interaction of TIA-1/TIAR with West Nile and Dengue Virus Products in Infected Cells Interferes with Stress Granule Formation and Processing Body Assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 9041–9046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Hu, Z.; Fan, S.; Zhang, Q.; Zhong, Y.; Guo, D.; Qin, Y.; Chen, M. Picornavirus 2A Protease Regulates Stress Granule Formation to Facilitate Viral Translation. PLoS Pathog. 2018, 14, e1006901. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Lin, L.; Si, X.; Wang, T.; Zhong, X.; Tong, L.; Luan, Y.; Chen, Y.; Li, X.; et al. Protease 2A Induces Stress Granule Formation during Coxsackievirus B3 and Enterovirus 71 Infections. Virol. J. 2014, 11, 192. [Google Scholar] [CrossRef] [Green Version]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clément, J.-F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 Ribonucleoproteins Prevent Formation of Stress Granules but Favour Encapsidation of HIV-1 Genomic RNA. J. Cell Sci. 2010, 123, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Legros, S.; Boxus, M.; Gatot, J.S.; Van Lint, C.; Kruys, V.; Kettmann, R.; Twizere, J.C.; Dequiedt, F. The HTLV-1 Tax Protein Inhibits Formation of Stress Granules by Interacting with Histone Deacetylase 6. Oncogene 2011, 30, 4050–4062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Ndongwe, T.P.; Puray-Chavez, M.; Casey, M.C.; Izumi, T.; Pathak, V.K.; Tedbury, P.R.; Sarafianos, S.G. Effect of P-Body Component Mov10 on HCV Virus Production and Infectivity. FASEB J. 2020, 34, 9433–9449. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, R.; Mukherjee, A.; Patra, U.; Chawla-Sarkar, M. Rotavirus Disrupts Cytoplasmic P Bodies during Infection. Virus Res. 2015, 210, 344–354. [Google Scholar] [CrossRef]
- Dougherty, J.D.; White, J.P.; Lloyd, R.E. Poliovirus-Mediated Disruption of Cytoplasmic Processing Bodies. J. Virol. 2011, 85, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Kesari, A.S.; Heintz, V.J.; Poudyal, S.; Miller, A.S.; Kuhn, R.J.; LaCount, D.J. Zika Virus NS5 Localizes at Centrosomes during Cell Division. Virology 2020, 541, 52–62. [Google Scholar] [CrossRef]
- Afonso, P.V.; Zamborlini, A.; Saïb, A.; Mahieux, R. Centrosome and Retroviruses: The Dangerous Liaisons. Retrovirology 2007, 4, 27. [Google Scholar] [CrossRef] [Green Version]
- Peloponese, J.-M.; Haller, K.; Miyazato, A.; Jeang, K.-T. Abnormal Centrosome Amplification in Cells through the Targeting of Ran-Binding Protein-1 by the Human T Cell Leukemia Virus Type-1 Tax Oncoprotein. Proc. Natl. Acad. Sci. USA 2005, 102, 18974–18979. [Google Scholar] [CrossRef] [Green Version]
- Davis, L.E.; DeBiasi, R.; Goade, D.E.; Haaland, K.Y.; Harrington, J.A.; Harnar, J.B.; Pergam, S.A.; King, M.K.; DeMasters, B.K.; Tyler, K.L. West Nile Virus Neuroinvasive Disease. Ann. Neurol. 2006, 60, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Misra, U.K.; Kalita, J. Overview: Japanese Encephalitis. Prog. Neurobiol. 2010, 91, 108–120. [Google Scholar] [CrossRef]
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.A.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.-Y.; Vasilakis, N. Zika Virus: History, Emergence, Biology, and Prospects for Control. Antivir. Res. 2016, 130, 69–80. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Gresh, L.; Halloran, M.E.; Mercado, J.C.; Kuan, G.; Gordon, A.; Balmaseda, A.; Harris, E. Antibody-Dependent Enhancement of Severe Dengue Disease in Humans. Science 2017, 358, 929–932. [Google Scholar] [CrossRef] [Green Version]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-Reacting Antibodies Enhance Dengue Virus Infection in Humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [Green Version]
- Bardina, S.V.; Bunduc, P.; Tripathi, S.; Duehr, J.; Frere, J.J.; Brown, J.A.; Nachbagauer, R.; Foster, G.A.; Krysztof, D.; Tortorella, D.; et al. Enhancement of Zika Virus Pathogenesis by Preexisting Antiflavivirus Immunity. Science 2017, 356, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Paul, L.M.; Carlin, E.R.; Jenkins, M.M.; Tan, A.L.; Barcellona, C.M.; Nicholson, C.O.; Michael, S.F.; Isern, S. Dengue Virus Antibodies Enhance Zika Virus Infection. Clin. Transl. Immunol. 2016, 5, e117. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Supasa, P.; Wongwiwat, W.; Rouvinski, A.; Barba-Spaeth, G.; Duangchinda, T.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Malasit, P.; Rey, F.A.; et al. Dengue Virus Sero-Cross-Reactivity Drives Antibody-Dependent Enhancement of Infection with Zika Virus. Nat. Immunol. 2016, 17, 1102–1108. [Google Scholar] [CrossRef]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The Endoplasmic Reticulum Provides the Membrane Platform for Biogenesis of the Flavivirus Replication Complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [Green Version]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Bühler, S.; Bartenschlager, R. The Non-Structural Protein 4A of Dengue Virus Is an Integral Membrane Protein Inducing Membrane Alterations in a 2K-Regulated Manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef] [Green Version]
- Kaufusi, P.H.; Kelley, J.F.; Yanagihara, R.; Nerurkar, V.R. Induction of Endoplasmic Reticulum-Derived Replication-Competent Membrane Structures by West Nile Virus Non-Structural Protein 4B. PLoS ONE 2014, 9, e84040. [Google Scholar] [CrossRef] [Green Version]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated Cleavages at the West Nile Virus NS4A-2K-NS4B Junctions Play a Major Role in Rearranging Cytoplasmic Membranes and Golgi Trafficking of the NS4A Protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [Green Version]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 Structures Reveal Surfaces for Associations with Membranes and the Immune System. Science 2014, 343, 881–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Okabayashi, T.; Yamashita, T.; Zhao, Z.; Wakita, T.; Yasui, K.; Hasebe, F.; Tadano, M.; Konishi, E.; Moriishi, K.; et al. Nuclear Localization of Japanese Encephalitis Virus Core Protein Enhances Viral Replication. J. Virol. 2005, 79, 3448–3458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogatyreva, N.S.; Finkelstein, A.V.; Galzitskaya, O.V. Trend of Amino Acid Composition of Proteins of Different Taxa. J. Bioinform. Comput. Biol. 2006, 4, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.-P.; Zhang, X.; Han, P.; Arora, N.; Anders, R.F.; Norton, R.S. Abundance of Intrinsically Unstructured Proteins in P. Falciparum and Other Apicomplexan Parasite Proteomes. Mol. Biochem. Parasitol. 2006, 150, 256–267. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically Disordered Protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Schad, E.; Tompa, P.; Hegyi, H. The Relationship between Proteome Size, Structural Disorder and Organism Complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins Struct. Funct. Genet. 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Pushker, R.; Mooney, C.; Davey, N.E.; Jacqué, J.-M.; Shields, D.C. Marked Variability in the Extent of Protein Disorder within and between Viral Families. PLoS ONE 2013, 8, e60724. [Google Scholar] [CrossRef] [Green Version]
- Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of Spontaneous Mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [CrossRef] [PubMed]
- Gromowski, G.D.; Firestone, C.-Y.; Whitehead, S.S. Genetic Determinants of Japanese Encephalitis Virus Vaccine Strain SA14-14-2 That Govern Attenuation of Virulence in Mice. J. Virol. 2015, 89, 6328–6337. [Google Scholar] [CrossRef] [Green Version]
- dos Santos, C.N.; Post, P.R.; Carvalho, R.; Ferreira, I.I.; Rice, C.M.; Galler, R. Complete Nucleotide Sequence of Yellow Fever Virus Vaccine Strains 17DD and 17D-213. Virus Res. 1995, 35, 35–41. [Google Scholar] [CrossRef]
- Treiber, D.K.; Williamson, J.R. Beyond Kinetic Traps in RNA Folding. Curr. Opin. Struct. Biol. 2001, 11, 309–314. [Google Scholar] [CrossRef]
- Bertrand, E.L.; Rossi, J.J. Facilitation of Hammerhead Ribozyme Catalysis by the Nucleocapsid Protein of HIV-1 and the Heterogeneous Nuclear Ribonucleoprotein A1. EMBO J. 1994, 13, 2904–2912. [Google Scholar] [CrossRef]
- Darlix, J.-L.; Lapadat-Tapolsky, M.; de Rocquigny, H.; Roques, B.P. First Glimpses at Structure-Function Relationships of the Nucleocapsid Protein of Retroviruses. J. Mol. Biol. 1995, 254, 523–537. [Google Scholar] [CrossRef]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-Acid-Chaperone Activity of Retroviral Nucleocapsid Proteins: Significance for Viral Replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef]
- Tompa, P.; Csermely, P. The Role of Structural Disorder in the Function of RNA and Protein Chaperones. FASEB J. 2004, 18, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Bahadur, R.P. A Structural Perspective of RNA Recognition by Intrinsically Disordered Proteins. Cell. Mol. Life Sci. 2016, 73, 4075–4084. [Google Scholar] [CrossRef]
- Poole, A.M.; Jeffares, D.C.; Penny, D. The Path from the RNA World. J. Mol. Evol. 1998, 46, 1–17. [Google Scholar] [CrossRef]
- Frankel, A.D.; Young, J.A.T. HIV-1: Fifteen Proteins and an RNA. Annu. Rev. Biochem. 1998, 67, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Summers, M.F.; Henderson, L.E.; Chance, M.R.; South, T.L.; Blake, P.R.; Perez-Alvarado, G.; Bess, J.W.; Sowder, R.C.; Arthur, L.O.; Sagi, I.; et al. Nucleocapsid Zinc Fingers Detected in Retroviruses: EXAFS Studies of Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein from HIV-1. Protein Sci. 1992, 1, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morellet, N.; Jullian, N.; De Rocquigny, H.; Maigret, B.; Darlix, J.L.; Roques, B.P. Determination of the Structure of the Nucleocapsid Protein NCp7 from the Human Immunodeficiency Virus Type 1 by 1H NMR. EMBO J. 1992, 11, 3059–3065. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Mizianty, M.J.; Kurgan, L.; Uversky, V.N. Protein Intrinsic Disorder as a Flexible Armor and a Weapon of HIV-1. Cell. Mol. Life Sci. 2012, 69, 1211–1259. [Google Scholar] [CrossRef]
- Carteau, S.; Batson, S.C.; Poljak, L.; Mouscadet, J.-F.O.; Darlix, J.-L. Human Immunodeficiency Virus Type 1 Nucleocapsid Protein Specifically Stimulates Mg2+-Dependent DNA Integration In Vitro. J. Virol. 1997, 71, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Khorchid, A.; Wang, J.; Parniak, M.A.; Darlix, J.L.; Wainberg, M.A.; Kleiman, L. Effect of Mutations in the Nucleocapsid Protein (NCp7) upon Pr160(Gag-Pol) and TRNA(Lys) Incorporation into Human Immunodeficiency Virus Type 1. J. Virol. 1997, 71, 4378–4384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Henderson, L.E.; Bess, J.; Kane, B.; Levin, J.G. Human Immunodeficiency Virus Type 1 Nucleocapsid Protein Promotes Efficient Strand Transfer and Specific Viral DNA Synthesis by Inhibiting TAR-Dependent Self-Priming from Minus-Strand Strong-Stop DNA. J. Virol. 1997, 71, 5178–5188. [Google Scholar] [CrossRef] [Green Version]
- Cameron, C.E.; Ghosh, M.; Le Grice, S.F.J.; Benkovic, S.J. Mutations in HIV Reverse Transcriptase Which Alter RNase H Activity and Decrease Strand Transfer Efficiency Are Suppressed by HIV Nucleocapsid Protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6700–6705. [Google Scholar] [CrossRef] [Green Version]
- Cristofari, G. The Hepatitis C Virus Core Protein Is a Potent Nucleic Acid Chaperone That Directs Dimerization of the Viral (+) Strand RNA in Vitro. Nucleic Acids Res. 2004, 32, 2623–2631. [Google Scholar] [CrossRef] [Green Version]
- Ivanyi-Nagy, R.; Kanevsky, I.; Gabus, C.; Lavergne, J.-P.; Ficheux, D.; Penin, F.; Fosse, P.; Darlix, J.-L. Analysis of Hepatitis C Virus RNA Dimerization and Core-RNA Interactions. Nucleic Acids Res. 2006, 34, 2618–2633. [Google Scholar] [CrossRef]
- Ivanyi-Nagy, R.; Lavergne, J.-P.; Gabus, C.; Ficheux, D.; Darlix, J.-L. RNA Chaperoning and Intrinsic Disorder in the Core Proteins of Flaviviridae. Nucleic Acids Res. 2008, 36, 712–725. [Google Scholar] [CrossRef]
- Pong, W.L.; Huang, Z.S.; Teoh, P.G.; Wang, C.C.; Wu, H.N. RNA Binding Property and RNA Chaperone Activity of Dengue Virus Core Protein and Other Viral RNA-Interacting Proteins. FEBS Lett. 2011, 585, 2575–2581. [Google Scholar] [CrossRef] [Green Version]
- Davey, J.; Colman, A.; Dimmock, N.J. Location of Influenza Virus M, NP and NS1 Proteins in Microinjected Cells. J. Gen. Virol. 1985, 66, 2319–2334. [Google Scholar] [CrossRef]
- Michienzi, A.; Cagnon, L.; Bahner, I.; Rossi, J.J. Ribozyme-Mediated Inhibition of HIV 1 Suggests Nucleolar Trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 2000, 97, 8955–8960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonntag, F.; Schmidt, K.; Kleinschmidt, J.A. A Viral Assembly Factor Promotes AAV2 Capsid Formation in the Nucleolus. Proc. Natl. Acad. Sci. USA 2010, 107, 10220–10225. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, Y.; Mori, Y.; Abe, T.; Yamashita, T.; Okamoto, T.; Ichimura, T.; Moriishi, K.; Matsuura, Y. Nucleolar Protein B23 Interacts with Japanese Encephalitis Virus Core Protein and Participates in Viral Replication. Microbiol. Immunol. 2006, 50, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.T.; Ma, L.; Burgner, J.W.; Groesch, T.D.; Post, C.B.; Kuhn, R.J. Flavivirus Capsid Is a Dimeric Alpha-Helical Protein. J. Virol. 2003, 77, 7143–7149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, T.Y.; Fibriansah, G.; Lok, S.-M. Capsid Protein Is Central to the Birth of Flavivirus Particles. PLoS Pathog. 2020, 16, e1008542. [Google Scholar] [CrossRef]
- Kiermayr, S.; Kofler, R.M.; Mandl, C.W.; Messner, P.; Heinz, F.X. Isolation of Capsid Protein Dimers from the Tick-Borne Encephalitis Flavivirus and in Vitro Assembly of Capsid-like Particles. J. Virol. 2004, 78, 8078–8084. [Google Scholar] [CrossRef] [Green Version]
- Poonsiri, T.; Wright, G.S.A.; Solomon, T.; Antonyuk, S.V. Crystal Structure of the Japanese Encephalitis Virus Capsid Protein. Viruses 2019, 11, 623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, T.Y.; Fibriansah, G.; Kostyuchenko, V.A.; Ng, T.-S.; Lim, X.-X.; Zhang, S.; Lim, X.-N.; Wang, J.; Shi, J.; Morais, M.C.; et al. Capsid Protein Structure in Zika Virus Reveals the Flavivirus Assembly Process. Nat. Commun. 2020, 11, 895. [Google Scholar] [CrossRef]
- Roby, J.A.; Setoh, Y.X.; Hall, R.A.; Khromykh, A.A. Post-Translational Regulation and Modifications of Flavivirus Structural Proteins. J. Gen. Virol. 2015, 96, 1551–1569. [Google Scholar] [CrossRef]
- Oliveira, E.R.A.; Mohana-Borges, R.; de Alencastro, R.B.; Horta, B.A.C. The Flavivirus Capsid Protein: Structure, Function and Perspectives towards Drug Design. Virus Res. 2017, 227, 115–123. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Westaway, E.G. RNA Binding Properties of Core Protein of the Flavivirus Kunjin. Arch. Virol. 1996, 141, 685–699. [Google Scholar] [CrossRef]
- Samsa, M.M.; Mondotte, J.A.; Caramelo, J.J.; Gamarnik, A.V. Uncoupling Cis-Acting RNA Elements from Coding Sequences Revealed a Requirement of the N-Terminal Region of Dengue Virus Capsid Protein in Virus Particle Formation. J. Virol. 2012, 86, 1046–1058. [Google Scholar] [CrossRef] [Green Version]
- Kofler, R.M.; Heinz, F.X.; Mandl, C.W. Capsid Protein C of Tick-Borne Encephalitis Virus Tolerates Large Internal Deletions and Is a Favorable Target for Attenuation of Virulence. J. Virol. 2002, 76, 3534–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlick, P.; Taucher, C.; Schittl, B.; Tran, J.L.; Kofler, R.M.; Schueler, W.; von Gabain, A.; Meinke, A.; Mandl, C.W. Helices A2 and A3 of West Nile Virus Capsid Protein Are Dispensable for Assembly of Infectious Virions. J. Virol. 2009, 83, 5581–5591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slomnicki, L.P.; Chung, D.H.; Parker, A.; Hermann, T.; Boyd, N.L.; Hetman, M. Ribosomal Stress and Tp53-Mediated Neuronal Apoptosis in Response to Capsid Protein of the Zika Virus. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Anderson, R.; Hobman, T.C. The Capsid-Binding Nucleolar Helicase DDX56 Is Important for Infectivity of West Nile Virus. J. Virol. 2011, 85, 5571–5580. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Goto, S.; Ishimura, M.; Amanuma, M.; Hara, Y.; Suzuki, R.; Katoh, K.; Morita, E. Functional Correlation between Subcellular Localizations of Japanese Encephalitis Virus Capsid Protein and Virus Production. J. Virol. 2019, 93, e00612-19. [Google Scholar] [CrossRef]
- Tiwary, A.K.; Cecilia, D. Kinetics of the Association of Dengue Virus Capsid Protein with the Granular Component of Nucleolus. Virology 2017, 502, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, M.; Lorinczi, M.; Rijnbrand, R.; Lemon, S.M.; Watowich, S.J. Self-Assembly of Nucleocapsid-Like Particles from Recombinant Hepatitis C Virus Core Protein. J. Virol. 2001, 75, 2119–2129. [Google Scholar] [CrossRef] [Green Version]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP–Caprin1–USP10 Complexes Mediate Stress Granule Condensation and Associate with 40S Subunits. J. Cell Biol. 2016, 212, e201508028. [Google Scholar] [CrossRef] [Green Version]
- Courtney, S.C.; Scherbik, S.V.; Stockman, B.M.; Brinton, M.A. West Nile Virus Infections Suppress Early Viral RNA Synthesis and Avoid Inducing the Cell Stress Granule Response. J. Virol. 2012, 86, 3647–3657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonenfant, G.; Williams, N.; Netzband, R.; Schwarz, M.C.; Evans, M.J.; Pager, C.T. Zika Virus Subverts Stress Granules To Promote and Restrict Viral Gene Expression. J. Virol. 2019, 93, 22. [Google Scholar] [CrossRef] [Green Version]
- Amorim, R.; Temzi, A.; Griffin, B.D.; Mouland, A.J. Zika Virus Inhibits EIF2α-Dependent Stress Granule Assembly. PLoS Negl. Trop. Dis. 2017, 11, e0005775. [Google Scholar] [CrossRef] [Green Version]
- Hou, S.; Kumar, A.; Xu, Z.; Airo, A.M.; Stryapunina, I.; Wong, C.P.; Branton, W.; Tchesnokov, E.; Götte, M.; Power, C.; et al. Zika Virus Hijacks Stress Granule Proteins and Modulates the Host Stress Response. J. Virol. 2017, 91, e00474-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blázquez, A.-B.; Martín-Acebes, M.A.; Poderoso, T.; Saiz, J.-C. Relevance of Oxidative Stress in Inhibition of EIF2 Alpha Phosphorylation and Stress Granules Formation during Usutu Virus Infection. PLOS Negl. Trop. Dis. 2021, 15, e0009072. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Courtney, S.C.; Brinton, M.A. Arsenite-Induced Stress Granule Formation Is Inhibited by Elevated Levels of Reduced Glutathione in West Nile Virus-Infected Cells. PLoS Pathog. 2017, 13, e1006240. [Google Scholar] [CrossRef]
- Bidet, K.; Dadlani, D.; Garcia-Blanco, M.A. G3BP1, G3BP2 and CAPRIN1 Are Required for Translation of Interferon Stimulated MRNAs and Are Targeted by a Dengue Virus Non-Coding RNA. PLOS Pathog. 2014, 10, e1004242. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.S.-Y.; Sze, L.; Lam, K.-P. The Stress Granule Protein G3BP1 Binds Viral DsRNA and RIG-I to Enhance Interferon-β Response. J. Biol. Chem. 2019, 294, 6430–6438. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Ru, Y.; Ren, J.; Bai, J.; Wei, J.; Fu, S.; Liu, X.; Li, D.; Zheng, H. G3BP1 Inhibits RNA Virus Replication by Positively Regulating RIG-I-Mediated Cellular Antiviral Response. Cell Death Dis. 2019, 10, 946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Alam, U.; Willis, C.; Kennedy, D. Role of Chikungunya NsP3 in Regulating G3BP1 Activity, Stress Granule Formation and Drug Efficacy. Arch. Med. Res. 2021, 52, 48–57. [Google Scholar] [CrossRef]
- Fros, J.J.; Domeradzka, N.E.; Baggen, J.; Geertsema, C.; Flipse, J.; Vlak, J.M.; Pijlman, G.P. Chikungunya Virus NsP3 Blocks Stress Granule Assembly by Recruitment of G3BP into Cytoplasmic Foci. J. Virol. 2012, 86, 10873–10879. [Google Scholar] [CrossRef] [Green Version]
- Götte, B.; Utt, A.; Fragkoudis, R.; Merits, A.; McInerney, G.M. Sensitivity of Alphaviruses to G3BP Deletion Correlates with Efficiency of Replicase Polyprotein Processing. J. Virol. 2020, 94, e01681-19. [Google Scholar] [CrossRef] [PubMed]
- Alam, U.; Kennedy, D. G3BP1 and G3BP2 Regulate Translation of Interferon-Stimulated Genes: IFITM1, IFITM2 and IFITM3 in the Cancer Cell Line MCF7. Mol. Cell. Biochem. 2019, 459, 189–204. [Google Scholar] [CrossRef]
- Reineke, L.C.; Tsai, W.-C.; Jain, A.; Kaelber, J.T.; Jung, S.Y.; Lloyd, R.E. Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Masaki, T.; Matsunaga, S.; Takahashi, H.; Nakashima, K.; Kimura, Y.; Ito, M.; Matsuda, M.; Murayama, A.; Kato, T.; Hirano, H.; et al. Involvement of Hepatitis C Virus NS5A Hyperphosphorylation Mediated by Casein Kinase I- in Infectious Virus Production. J. Virol. 2014, 88, 7541–7555. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Hwang, J.; Sharma, S.D.; Hargittai, M.R.S.; Chen, Y.; Arnold, J.J.; Raney, K.D.; Cameron, C.E. Hepatitis C Virus Nonstructural Protein 5A (NS5A) Is an RNA-Binding Protein. J. Biol. Chem. 2005, 280, 36417–36428. [Google Scholar] [CrossRef] [Green Version]
- Morozova, O.V.; Tsekhanovskaya, N.A.; Maksimova, T.G.; Bachvalova, V.N.; Matveeva, V.A.; Kit, Y.Y. Phosphorylation of Tick-Borne Encephalitis Virus NS5 Protein. Virus Res. 1997, 49, 9–15. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Kenney, M.T.; Westaway, E.G. West Nile Virus Strain Kunjin NS5 Polymerase Is a Phosphoprotein Localized at the Cytoplasmic Site of Viral RNA Synthesis. J. Gen. Virol. 2007, 88, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.E.; Gorbalenya, A.E.; Rice, C.M. The NS5A/NS5 Proteins of Viruses from Three Genera of the Family Flaviviridae Are Phosphorylated by Associated Serine/Threonine Kinases. J. Virol. 1998, 72, 6199–6206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, M.; Zhang, L.; Ramachandra, M.; Kusukawa, J.; Ebner, K.E.; Padmanabhan, R. Association between NS3 and NS5 Proteins of Dengue Virus Type 2 in the Putative RNA Replicase Is Linked to Differential Phosphorylation of NS5. J. Biol. Chem. 1995, 270, 19100–19106. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Ansari, I.H.; Striker, R. The Flaviviral Methyltransferase Is a Substrate of Casein Kinase 1. Virus Res. 2009, 141, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keating, J.A.; Bhattacharya, D.; Lim, P.-Y.; Falk, S.; Weisblum, B.; Bernard, K.A.; Sharma, M.; Kuhn, R.J.; Striker, R. West Nile Virus Methyltransferase Domain Interacts with Protein Kinase G. Virol. J. 2013, 10, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iakoucheva, L.M. The Importance of Intrinsic Disorder for Protein Phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Kosako, H.; Yoshimura, S.H. Quantitative Proteomics Indicate a Strong Correlation of Mitotic Phospho-/Dephosphorylation with Non-Structured Regions of Substrates. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2020, 1868, 140295. [Google Scholar] [CrossRef]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to MTORC1 Signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.T.; Smith, J.; Chen, B.-C.; Schmidt, H.; Rasoloson, D.; Paix, A.; Lambrus, B.G.; Calidas, D.; Betzig, E.; Seydoux, G. Regulation of RNA Granule Dynamics by Phosphorylation of Serine-Rich, Intrinsically Disordered Proteins in C. Elegans. eLife 2014, 3, e04591. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Conicella, A.E.; Schmidt, H.B.; Martin, E.W.; Rhoads, S.N.; Reeb, A.N.; Nourse, A.; Ramirez Montero, D.; Ryan, V.H.; Rohatgi, R.; et al. A Single N-terminal Phosphomimic Disrupts TDP-43 Polymerization, Phase Separation, and RNA Splicing. EMBO J. 2018, 37, e97452. [Google Scholar] [CrossRef]
- Greig, J.A.; Nguyen, T.A.; Lee, M.; Holehouse, A.S.; Posey, A.E.; Pappu, R.V.; Jedd, G. Arginine-Enriched Mixed-Charge Domains Provide Cohesion for Nuclear Speckle Condensation. Mol. Cell 2020, 77, 1237–1250. [Google Scholar] [CrossRef]
- Das, S.; Eisen, A.; Lin, Y.-H.; Chan, H.S. A Lattice Model of Charge-Pattern-Dependent Polyampholyte Phase Separation. J. Phys. Chem. B 2018, 122, 5418–5431. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-H.; Chan, H.S. Phase Separation and Single-Chain Compactness of Charged Disordered Proteins Are Strongly Correlated. Biophys. J. 2017, 112, 2043–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-H.; Song, J.; Forman-Kay, J.D.; Chan, H.S. Random-Phase-Approximation Theory for Sequence-Dependent, Biologically Functional Liquid-Liquid Phase Separation of Intrinsically Disordered Proteins. J. Mol. Liq. 2017, 228, 176–193. [Google Scholar] [CrossRef] [Green Version]
- Castelnovo, M.; Joanny, J.F. Phase Diagram of Diblock Polyampholyte Solutions. Macromolecules 2002, 35, 4531–4538. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Cui, Y.; Han, X.; Hu, W.; Sun, M.; Zhang, Y.; Wang, P.-H.; Song, G.; Chen, W.; Lou, J. Liquid–Liquid Phase Separation by SARS-CoV-2 Nucleocapsid Protein and RNA. Cell Res. 2020, 30, 1143–1145. [Google Scholar] [CrossRef]
- Carlson, C.R.; Asfaha, J.B.; Ghent, C.M.; Howard, C.J.; Hartooni, N.; Safari, M.; Frankel, A.D.; Morgan, D.O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for Its Dual Functions. Mol. Cell 2020, 80, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Savastano, A.; Ibáñez de Opakua, A.; Rankovic, M.; Zweckstetter, M. Nucleocapsid Protein of SARS-CoV-2 Phase Separates into RNA-Rich Polymerase-Containing Condensates. Nat. Commun. 2020, 11, 6041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yu, Y.; Sun, L.-M.; Xing, J.-Q.; Li, T.; Zhu, Y.; Wang, M.; Yu, Y.; Xue, W.; Xia, T.; et al. GCG Inhibits SARS-CoV-2 Replication by Disrupting the Liquid Phase Condensation of Its Nucleocapsid Protein. Nat. Commun. 2021, 12, 2114. [Google Scholar] [CrossRef]
- Guseva, S.; Milles, S.; Jensen, M.R.; Salvi, N.; Kleman, J.-P.; Maurin, D.; Ruigrok, R.W.H.; Blackledge, M. Measles Virus Nucleo- and Phosphoproteins Form Liquid-like Phase-Separated Compartments that Promote Nucleocapsid Assembly. Sci. Adv. 2020, 6, eaaz7095. [Google Scholar] [CrossRef] [Green Version]
- Nevers, Q.; Albertini, A.A.; Lagaudrière-Gesbert, C.; Gaudin, Y. Negri Bodies and Other Virus Membrane-Less Replication Compartments. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118831. [Google Scholar] [CrossRef]
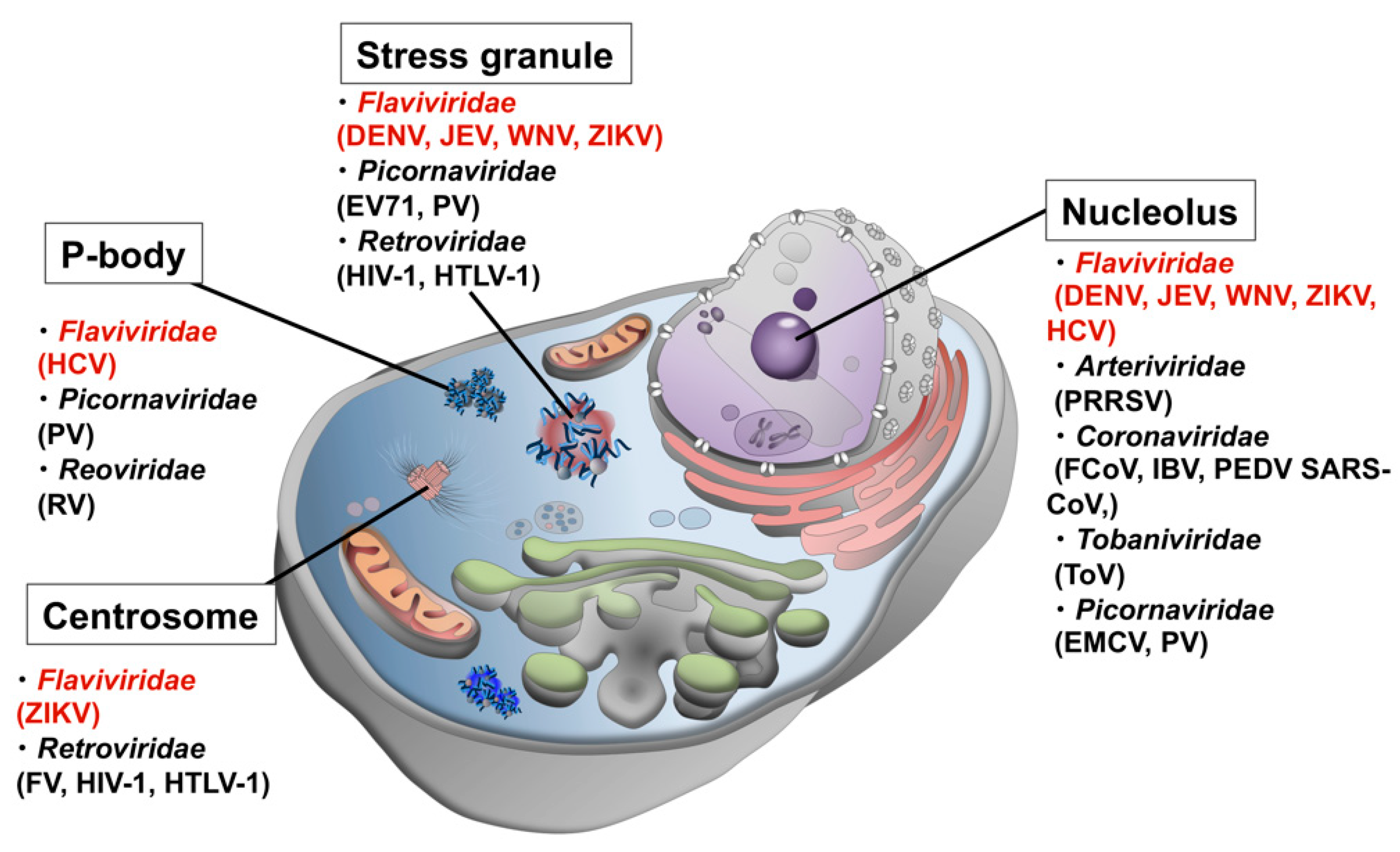


| MLOs | Viral Family | Viral Protein (Virus) | References |
|---|---|---|---|
| Nucleolus | Flaviviridae | Core (JEV, DENV, WNV, ZIKV) | [4] |
| NS5 (DENV), NS5B (HCV) | [5,6] | ||
| Nidovirales (Tobaniviridae, Arteriviridae, and Coronaviridae) | N (ToV, PEDV, SARS-CoV, IBV, PRRSV) | [7,8] | |
| NSP3B (FCoV, SARS-CoV) | [9,10] | ||
| Picornaviridae | 2A (EMCV) | [11] | |
| SG | Flaviviridae | Core (JEV) | [12] |
| NSs (WNV, DENV) | [13] | ||
| Picornaviridae | 2A (PV, EV71) | [14,15] | |
| Retroviridae | Gag (HIV-1) | [16] | |
| Tax (HTLV-1) | [17] | ||
| P-body | Flaviviridae | Core, NS5A? (HCV) | [18] |
| Reoviridae | NSP1 (RV) | [19] | |
| Picornaviridae | 3C (PV) | [20] | |
| Centrosome | Flaviviridae | NS5 (ZIKV) | [21] |
| Retroviridae | Gag (HIV-1, FV) | [22] | |
| Tax (HTLV-1) | [23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, A.; Shofa, M.; Ode, H.; Yumiya, M.; Hirano, J.; Okamoto, T.; Yoshimura, S.H. How Do Flaviviruses Hijack Host Cell Functions by Phase Separation? Viruses 2021, 13, 1479. https://doi.org/10.3390/v13081479
Saito A, Shofa M, Ode H, Yumiya M, Hirano J, Okamoto T, Yoshimura SH. How Do Flaviviruses Hijack Host Cell Functions by Phase Separation? Viruses. 2021; 13(8):1479. https://doi.org/10.3390/v13081479
Chicago/Turabian StyleSaito, Akatsuki, Maya Shofa, Hirotaka Ode, Maho Yumiya, Junki Hirano, Toru Okamoto, and Shige H. Yoshimura. 2021. "How Do Flaviviruses Hijack Host Cell Functions by Phase Separation?" Viruses 13, no. 8: 1479. https://doi.org/10.3390/v13081479
APA StyleSaito, A., Shofa, M., Ode, H., Yumiya, M., Hirano, J., Okamoto, T., & Yoshimura, S. H. (2021). How Do Flaviviruses Hijack Host Cell Functions by Phase Separation? Viruses, 13(8), 1479. https://doi.org/10.3390/v13081479






