Risk Assessment of Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis Patients during 1 Year of Ocrelizumab Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants and Samples Collection
2.2. Virological Investigations
2.3. Serological Investigations
2.4. Statistical Analysis
3. Results
3.1. JCPyV DNA in Urine and Plasma at Different Follow-Up Times
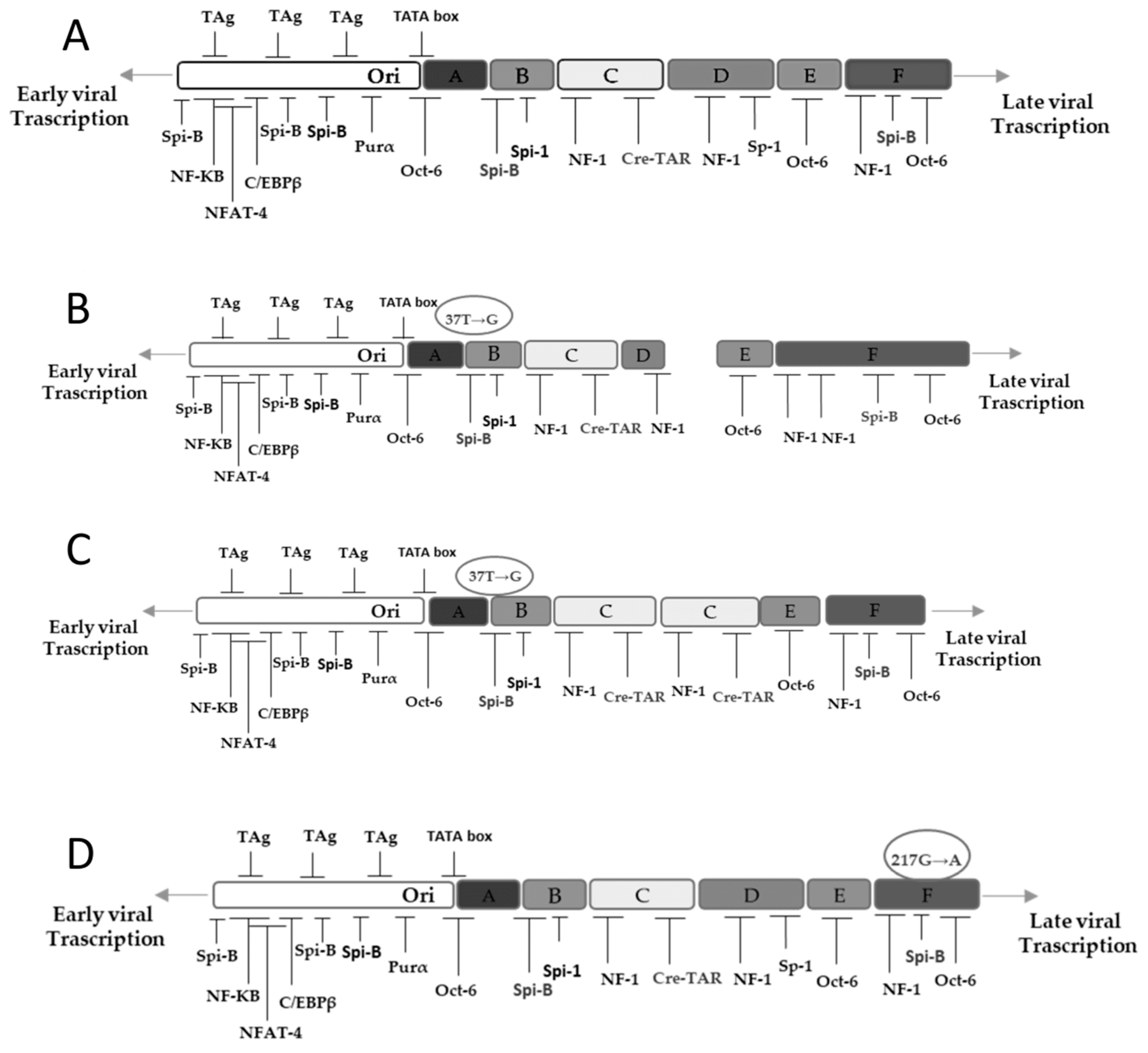
3.2. NCCR and VP1 Analysis
3.3. Phylogenetic Analysis
3.4. Assessment of Immunoglobulins and Anti-JCPyV Index in MS Patients
3.5. Combined Monitoring of the Serostatus and Its Relationship to Viral DNA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aksamit, A.J.; Major, E.O.; Ghatak, N.R.; Sidhu, G.S.; Parisi, J.E.; Guccion, J.G. Diagnosis of progressive multifocal leukoencephalopathy by brain biopsy with biotin labeled DNA: DNA in situ hybridization. J. Neuropathol. Exp. Neurol. 1987, 46, 556–566. [Google Scholar] [CrossRef]
- Houff, S.A.; Katz, D.; Kufta, C.V.; Major, E.O. A rapid method for in situ hybridization for viral DNA in brain biopsies from patients with AIDS. AIDS 1989, 3, 843–845. [Google Scholar] [CrossRef]
- Padgett, B.; ZuRhein, G.; Walker, D.; Eckroade, R.; Dessel, B. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 297, 1257–1260. [Google Scholar] [CrossRef]
- Assetta, B.; Atwood, W.J. The biology of JC polyomavirus. Biol. Chem. 2017, 398, 839–855. [Google Scholar] [CrossRef]
- Pietropaolo, V.; Prezioso, C.; Bagnato, F.; Antonelli, G. John Cunningham virus: An overview on biology and disease of the etiological agent of the progressive multifocal leukoencephalopathy. New Microbiol. 2018, 41, 179–186. [Google Scholar] [PubMed]
- McIlroy, D.; Halary, F.; Bressollette-Bodin, C. Intra-patient viral evolution in polyomavirus-related diseases. Philos. Trans. R. Soc. B 2019, 374, 20180301. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.N.; Khalili, K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J. Virol. 1995, 69, 5843–5848. [Google Scholar] [CrossRef] [Green Version]
- Sadowska, B.; Barrucco, R.; Khalili, K.; Safak, M. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J. Virol. 2003, 77, 665–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romagnoli, L.; Sariyer, I.K.; Tung, J.; Feliciano, M.; Sawaya, B.E.; Del Valle, L.; Ferrante, P.; Khalili, K.; Safak, M.; White, M.K. Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter. Virology 2008, 375, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedele, C.G.; Ciardi, M.R.; Delia, S.; Contreras, G.; Perez, J.L.; De Oña, M.; Vidal, E.; Tenorio, A. Identical rearranged forms of JC polyomavirus transcriptional control region in plasma and cerebrospinal fluid of acquired immunodeficiency syndrome patients with progressive multifocal leukoencephalopathy. J. Neurovirol. 2003, 9, 551–558. [Google Scholar] [CrossRef]
- Yogo, Y.; Kitamura, T.; Sugimoto, C.; Ueki, T.; Aso, Y.; Hara, K.; Taguchi, F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J. Virol. 1990, 64, 3139–3143. [Google Scholar] [CrossRef] [Green Version]
- Åström, K.-E.; Mancall, E.L.; Richardson, J.E.P. Progressive multifocal leuko-encephalopathy. Brain 1958, 81, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Walker, D.L. Progressive multifocal leukoencephalopathy. Neurol. Clin. 1984, 2, 299–313. [Google Scholar] [CrossRef]
- Tavazzi, E.; White, M.K.; Khalili, K. Progressive multifocal leukoencephalopathy: Clinical and molecular aspects. Rev. Med. Virol. 2012, 22, 18–32. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.R.; Scott, G.; Albrecht, J.; Belman, A.L.; Tornatore, C.; Major, E.O. Progressive multifocal leukoencephalopathy in HIV-1-infected children. AIDS 1992, 6, 837–841. [Google Scholar] [CrossRef]
- Wollebo, H.S.; White, M.K.; Gordon, J.; Berger, J.R.; Khalili, K. Persistence and pathogenesis of the neurotropic polyomavirus JC. Ann. Neurol. 2015, 77, 560–570. [Google Scholar] [CrossRef]
- Morriss, M.C.; Rutstein, R.M.; Rudy, B.; Desrochers, C.; Hunter, J.V.; Zimmerman, R.A. Progressive multifocal leukoencephalopathy in an HIV-infected child. Neuroradiology 1997, 39, 142–144. [Google Scholar] [CrossRef]
- Jelcic, I.; Kempf, C.; Largey, F.; Planas, R.; Schippling, S.; Budka, H.; Sospedra, M.; Martin, R. Mechanisms of immune escape in central nervous system infection with neurotropic JC virus variant. Ann. Neurol. 2016, 79, 404–418. [Google Scholar] [CrossRef]
- Cinque, P.; Koralnik, I.J.; Clifford, D.B. The evolving face of human immunodeficiency virus-related progressive multifocal leukoencephalopathy: Defining a consensus terminology. J. Neurovirol. 2003, 9, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Wolbers, M.; Mueller, N.J.; Garzoni, C.; Du Pasquier, R.A.; Fux, C.A.; Vernazza, P.; Bernasconi, E.; Viscidi, R.; Battegay, M.; et al. JC virus-specific immune responses in human immunodeficiency virus type 1 patients with progressive multifocal leukoencephalopathy. J. Virol. 2009, 83, 4404–4411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garvey, L.; Winston, A.; Walsh, J.; Post, F.; Porter, K.; Gazzard, B.; Fisher, M.; Leen, C.; Pillay, D.; Hill, T.; et al. Antiretroviral therapy CNS penetration and HIV-1-associated CNS disease. Neurology 2011, 76, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shitrit, D.; Lev, N.; Bar-Gil-Shitrit, A.; Kramer, M.R. Progressive multifocal leukoencephalopathy in transplant recipients. Transpl. Int. 2005, 17, 658–665. [Google Scholar] [CrossRef]
- Hecht, J.H.; Glenn, O.A.; Wara, D.W.; Wu, Y.W. JC virus granule cell neuronopathy in a child with CD40 ligand deficiency. Pediatr. Neurol. 2007, 36, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Redfearn, A.; Pennie, R.A.; Mahony, J.B.; Dent, P.B. Progressive multifocal leukoencephalopathy in a child with immunodeficiency and hyperimmunoglobulinemia M. Pediatric Infect. Dis. J. 1993, 12, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Van Assche, G.; Van Ranst, M.; Sciot, R.; Dubois, B.; Vermeire, S.; Noman, M.; Verbeeck, J.; Geboes, K.; Robberecht, W.; Rutgeerts, P. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. N. Engl. J. Med. 2005, 353, 362–368. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, K.J.; Hogg, J.P. Progressive multifocal leukoencephalopathy in patients with multiple sclerosis. Curr. Opin. Neurol. 2013, 26, 318–323. [Google Scholar] [CrossRef]
- Prezioso, C.; Zingaropoli, M.A.; Iannetta, M.; Rodio, D.M.; Altieri, M.; Conte, A.; Vullo, V.; Ciardi, M.R.; Palamara, A.T.; Pietropaolo, V. Which is the best PML risk stratification strategy in natalizumab-treated patients affected by multiple sclerosis? Mult. Scler. Relat. Disord. 2020, 41, 102008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langer-Gould, A.; Atlas, S.W.; Green, A.J.; Bollen, A.W.; Pelletier, D. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N. Engl. J. Med. 2005, 353, 375–381. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, B.; Kappos, L. Natalizumab: Targeting α4-Integrins in multiple sclerosis. Neurodegener. Dis. 2008, 5, 16–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinschmidt-DeMasters, B.K.; Tyler, K.L. Progressive multifocal leukoencephalo-pathy complicating treatment with natalizumab and interferon beta-1a for multiplesclerosis. N. Engl. J. Med. 2005, 353, 369–374. [Google Scholar] [CrossRef] [Green Version]
- Bloomgren, G.; Richman, S.; Hotermans, C.; Subramanyam, M.; Goelz, S.; Natarajan, A.; Lee, S.; Plavina, T.; Scanlon, J.V.; Sandrock, A.; et al. Risk of natalizumab-associated progressive multifocal leukoencephalopathy. N. Engl. J. Med. 2012, 366, 1870–1880. [Google Scholar] [CrossRef]
- Carson, K.R.; Evens, A.M.; Richey, E.A.; Habermann, T.M.; Focosi, D.; Seymour, J.F.; Laubach, J.; Bawn, S.D.; Gordon, L.I.; Winter, J.N.; et al. Progressive multifocal leukoencephalopathy after rituximab therapy in HIV-negative patients: A report of 57 cases from the Research on Adverse Drug Events and Reports project. Blood 2009, 113, 4834–4840. [Google Scholar] [CrossRef] [Green Version]
- McGinley, M.P.; Moss, B.P.; Cohen, J.A. Safety of monoclonal antibodies for the treatment of multiple sclerosis. Expert Opin. Drug Saf. 2017, 16, 89–100. [Google Scholar] [CrossRef]
- Rudnicka, D.; Oszmiana, A.; Finch, D.K.; Strickland, I.; Schofield, D.J.; Lowe, D.C.; Sleeman, M.A.; Davis, D.M. Rituximab causes a polarization of B cells that augments its therapeutic function in NK-cell-mediated antibody-dependent cellular cytotoxicity. Blood 2013, 121, 4694–4702. [Google Scholar] [CrossRef] [PubMed]
- Duddy, M.; Bar-Or, A. B-cells in multiple sclerosis. Int. MS J. 2006, 13, 84–90. [Google Scholar]
- Raisch, D.W.; Rafi, J.A.; Chen, C.; Bennett, C.L. Detection of cases of progressive multifocal leukoencephalopathy associated with new biologicals and targeted cancer therapies from the FDA’s adverse event reporting system. Expert Opin. Drug Saf. 2016, 15, 1003–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baber, U.; Bouley, A.; Egnor, E.; Sloane, J.A. Anti-JC virus antibody index changes in rituximab-treated multiple sclerosis patients. J. Neurol. 2018, 265, 2342–2345. [Google Scholar] [CrossRef] [PubMed]
- Genentech. Ocrelizumab & PML. Available online: www.ocrelizumabinfo.com (accessed on 4 August 2021).
- Berger, J.R. Classifying PML risk with disease modifying therapies. Mult. Scler. Relat. Disord. 2017, 12, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef] [Green Version]
- Delbue, S.; Branchetti, E.; Boldorini, R.; Vago, L.; Zerbi, P.; Veggiani, C.; Tremolada, S.; Ferrante, P. Presence and expression of JCV early gene large T antigen in the brains of immunocompromised and immunocompetent individuals. J. Med. Virol. 2008, 80, 2147–2152. [Google Scholar] [CrossRef] [Green Version]
- Prezioso, C.; Scribano, D.; Rodio, D.M.; Ambrosi, C.; Trancassini, M.; Palamara, A.T.; Pietropaolo, V. COS-7-based model: Methodological approach to study John Cunningham virus replication cycle. Virol. J. 2018, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Flægstad, T.; Sundsfjord, A.; Arthur, R.R.; Pedersen, M.; Traavik, T.; Subramani, S. Amplification and sequencing of the control regions of BK and JC virus from human urine by polymerase chain reaction. Virology 1991, 180, 553–560. [Google Scholar] [CrossRef]
- Markowitz, R.B.; Thompson, H.C.; Mueller, J.F.; Cohen, J.A.; Dynan, W.S. Incidence of BK virus and JC virus viruria in human immunodeficiency virus-infected and uninfected subjects. J. Infect. Dis. 1993, 167, 13–20. [Google Scholar] [CrossRef]
- Jin, L.; Gibson, P.E.; Knowles, W.A.; Clewley, J.P. BK virus antigenic variants: Sequence analysis within the capsid VP1 epitope. J. Med. Virol. 1993, 39, 50–56. [Google Scholar] [CrossRef] [PubMed]
- ClustalW2–Multiple Sequence Alignment. Available online: http://www.ebi.ac.uk/Tools/msa/clustalw2/ (accessed on 4 August 2021).
- Jobes, D.V.; Friedlaender, J.S.; Mgone, C.S.; Agostini, H.T.; Koki, G.; Yanagihara, R.; Ng, T.C.N.; Chima, S.C.; Ryschkewitsch, C.F.; Stoner, G.L. New JC virus (JCV) genotypes from papua new guinea and micronesia (type 8 and type 2E) and evolutionary analysis of 32 complete JCV genomes. Arch. Virol. 2001, 146, 2097–2113. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, T.; Rempe, T.; Leypoldt, F.; Riedel, C.; Jansen, O.; Berg, D.; Deuschl, G. The spectrum of progressive multifocal leukoencephalopathy: A practical approach. Eur. J. Neurol. 2019, 26, 566-e41. [Google Scholar] [CrossRef] [PubMed]
- Plavina, T.; Subramanyam, M.; Bloomgren, G.; Richman, S.; Pace, A.; Lee, S.; Schlain, B.; Campagnolo, D.; Belachew, S.; Ticho, B. Anti-JC virus antibody levels in serum or plasma further define risk of natalizumab-associated progressive multifocal leukoencephalopathy. Ann. Neurol. 2014, 76, 802–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.; Plavina, T.; Castro, A.; Berman, M.; Jaiswal, D.; Rivas, S.; Schlain, B.; Subramanyam, M. A second-generation ELISA (STRATIFY JCV™ DxSelect™) for detection of JC virus antibodies in human serum and plasma to support progressive multifocal leukoencephalopathy risk stratification. J. Clin. Virol. 2013, 57, 141–146. [Google Scholar] [CrossRef]
- Schwab, N.; Schneider-Hohendorf, T.; Pignolet, B.; Breuer, J.; Gross, C.C.; Göbel, K.; Brassat, D.; Wiendl, H. Therapy with natalizumab is associated with high JCV seroconversion and rising JCV index values. Neurol. Neuroimmunol. Neuroinflammation 2016, 3, e195. [Google Scholar] [CrossRef] [Green Version]
- Farley, S.; Gottesman, M.H.; Friedman-Urevich, S.; Ye, J.; Shen, M.; Grueneberg, D.; Martone, L.; Calixte, R. Anti-John Cunningham virus antibody index levels in multiple sclerosis patients treated with rituximab, fingolimod, and dimethyl fumarate. Surg. Neurol. Int. 2019, 10, 59. [Google Scholar] [CrossRef]
- Williamson, E.; Dobrowolski, J. Impact of Ocrelizumab Treatment on PML Risk Biomarkers; Consortium of Multiple Sclerosis Centers: Nashville, TN, USA, 2018. [Google Scholar]
- Focosi, D.; Tuccori, M.; Maggi, F. Progressive multifocal leukoencephalopathy and anti-CD20 monoclonal antibodies: What do we know after 20 years of rituximab. Rev. Med. Virol. 2019, 29, e2077. [Google Scholar] [CrossRef]
- Chen, Y.; Bord, E.; Tompkins, T.; Miller, J.; Tan, C.S.; Kinkel, R.P.; Stein, M.C.; Viscidi, R.P.; Ngo, L.H.; Koralnik, I.J. Asymptomatic reactivation of JC virus in patients treated with natalizumab. N. Engl. J. Med. 2009, 361, 1067–1074. [Google Scholar] [CrossRef]
- White, M.K.; Safak, M.; Khalili, K. Regulation of gene expression in primate polyomaviruses. J. Virol. 2009, 83, 10846–10856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.U.; Tang, S.C.; Pater, M.M.; Pater, A. Glial and muscle embryonal carcinoma cell-specific independent regulation of expression of human JC virus early promoter by cyclic AMP response elements and adjacent nuclear factor 1 binding sites. J. Med. Virol. 1996, 49, 199–204. [Google Scholar] [CrossRef]
- Ciardi, M.R.; Zingaropoli, M.A.; Iannetta, M.; Prezioso, C.; Perri, V.; Pasculli, P.; Lichtner, M.; d’Ettorre, G.; Altieri, M.; Conte, A.; et al. JCPyV NCCR analysis in PML patients with different risk factors: Exploring common rearrangements as essential changes for neuropathogenesis. Virol. J. 2020, 17, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, L.J.; Moore, L.D.; Mirsky, M.M.; Major, E.O. JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes. J. Gen. Virol. 2012, 93, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Sunyaev, S.R.; Lugovskoy, A.; Simon, K.; Gorelik, L. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML). PLoS Genet. 2009, 5, e1000368. [Google Scholar] [CrossRef] [Green Version]
- Agostini, H.T.; Deckhut, A.; Jobes, D.V.; Girones, R.; Schlunck, G.; Prost, M.G.; Frias, C.; Pérez-Trallero, E.; Ryschkewitsch, C.F.; Stoner, G.L. Genotypes of JC virus in East, Central and Southwest Europe. J. Gen. Virol. 2001, 82, 1221–1331. [Google Scholar] [CrossRef] [Green Version]
- Anselmo, A.; Prezioso, C.; Saccà, F.A.; Di Lella, F.M.; Palmieri, G.; Tisone, G.; Pietropaolo, V.; Ciotti, M. Kidney graft failure induced by BKPyV replication despite a strong reduction of the immunosuppressive therapy. J. Med. Virol. 2019, 91, 1698–1701. [Google Scholar] [CrossRef]
- Prezioso, C.; Bianchi, M.; Obregon, F.; Ciotti, M.; Sarmati, L.; Andreoni, M.; Palamara, A.T.; Pascarella, S.; Moens, U.; Pietropaolo, V. Structural Analysis of Merkel Cell Polyomavirus (MCPyV) Viral Capsid Protein 1 (VP1) in HIV-1 Infected Individuals. Int. J. Mol. Sci. 2020, 21, 7998. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Jones, R.B.; Smith, R.M.; Alberici, F.; Kumaratne, D.S.; Burns, S.; Jayne, D.R. Rituximab-associated hypogammaglobulinemia: Incidence, predictors and outcomes in patients with multi-system autoimmune disease. J. Autoimmun. 2015, 57, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Prezioso, C.; Pietropaolo, V. An overview on human polyomaviruses biology and related diseases. Future Virol. 2019, 14, 487–501. [Google Scholar] [CrossRef]
- Major, E.O.; Yousry, T.A.; Clifford, D.B. Pathogenesis of progressive multifocal leukoencephalopathy and risks associated with treatments for multiple sclerosis: A decade of lessons learned. Lancet Neurol. 2018, 17, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Correia, I.; Jesus-Ribeiro, J.; Batista, S.; Martins, A.I.; Nunes, C.; Macário, M.C.; Cunha, L.; Sousa, L. Anti-JCV antibody serostatus and longitudinal evaluation in a Portuguese Multiple Sclerosis population. J. Clin. Neurosci. 2017, 45, 257–260. [Google Scholar] [CrossRef]
- Brosseau, M.S.G.; Stobbe, G.; Wundes, A. Natalizumab-related PML 2 weeks after negative anti-JCV antibody assay. Neurology 2016, 86, 484–486. [Google Scholar] [CrossRef]
| Features | Population | ||
|---|---|---|---|
| Patients, n | 42 | ||
| Sex, n (M/F) | M | F | |
| 18 (42.8%) | 24 (57.1%) | ||
| Mean age, years (SD) | 40.34 (± 9.01) | ||
| Median age, years (Range) | 40.5 (56.5–22.0) | ||
| Diagnosis, n | RRMS | ||
| 42 | |||
| Pre-treatments, n | Naive | Natalizumab | Others * |
| 19 | 4 | 19 | |
| Features | T0 | T2 | T4 |
|---|---|---|---|
| Positive viruria, n | 34/42 (81%) | 34/42 (81%) | 35/42 (83%) |
| Range, copies/mL | 1.1 × 106 4 × 104 | 5.8 × 107 1.2 × 105 | 9 × 109 1 × 106 |
| Mean viruria, copies/mL | 7.6 × 105 | 5.3 × 106 | 3.8 × 107 |
| Positive viremia, n | 0/42 (0%) | 0/42 (0%) | 3/42 (7%) |
| Range, copies/mL | 0 | 0 | 2.0 × 103 1.5 × 102 |
| Mean viremia, copies/mL | 0 | 0 | 6.5 × 102 |
| Mean T0 (SD) | Mean T2 (SD) | Mean T4 (SD) | F | |
|---|---|---|---|---|
| IgG | 960.08 (±202.22) | 964.22 (±180.05) | 978.14 (±191.09) | p > 0.05 |
| IgM | 127.95 (±53.50) | 98.19 (±55.57) | 75.04 (±41.93) | p < 0.05 |
| Features | T0 | T2 | T4 | F |
|---|---|---|---|---|
| JCV index ≥ 1.5, n | 26 | 26 | 27 | |
| 0.9 < JCV index > 1.5, n | 1 | 1 | 1 | |
| JCV index ≤ 0.9, n | 15 | 15 | 14 | |
| Mean JCV index (SD) | 2.24 (±1.53) | 1.78 (±1.45) | 1.56 (±1.38) | p < 0.05 |
| T0 | T2 | T4 | |
|---|---|---|---|
| JCV index > 1.5, n | 26/42 | 26/42 | 27/42 |
| Positive viruria, n | 26/34 | 26/34 | 27/35 |
| Mean viruria, copies/mL | 1.0 × 106 | 9.0 × 106 | 8.8 × 107 |
| Positive viremia, n | 0 | 0 | 3 |
| Mean viruria, copies/mL | 1.5 × 103 | ||
| 0.9 < JCV < 1.5 | 1/42 | 1/42 | 1/42 |
| Positive viruria, n | 1/34 | 1/34 | 1/35 |
| Viruria load, copies/mL | 7.5 × 104 | 1.5 × 105 | 2.0 × 103 |
| Positive viremia, n | 0 | 0 | 0 |
| JCV ≤ 0.9 | 15/42 | 15/42 | 14/42 |
| Positive viruria, n | 15/34 | 15/34 | 14/35 |
| Mean viruria, copies/mL | 3.0 × 103 | 9.8 × 103 | 1.0 × 104 |
| Positive viremia, n | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prezioso, C.; Grimaldi, A.; Landi, D.; Nicoletti, C.G.; Brazzini, G.; Piacentini, F.; Passerini, S.; Limongi, D.; Ciotti, M.; Palamara, A.T.; et al. Risk Assessment of Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis Patients during 1 Year of Ocrelizumab Treatment. Viruses 2021, 13, 1684. https://doi.org/10.3390/v13091684
Prezioso C, Grimaldi A, Landi D, Nicoletti CG, Brazzini G, Piacentini F, Passerini S, Limongi D, Ciotti M, Palamara AT, et al. Risk Assessment of Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis Patients during 1 Year of Ocrelizumab Treatment. Viruses. 2021; 13(9):1684. https://doi.org/10.3390/v13091684
Chicago/Turabian StylePrezioso, Carla, Alfonso Grimaldi, Doriana Landi, Carolina Gabri Nicoletti, Gabriele Brazzini, Francesca Piacentini, Sara Passerini, Dolores Limongi, Marco Ciotti, Anna Teresa Palamara, and et al. 2021. "Risk Assessment of Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis Patients during 1 Year of Ocrelizumab Treatment" Viruses 13, no. 9: 1684. https://doi.org/10.3390/v13091684
APA StylePrezioso, C., Grimaldi, A., Landi, D., Nicoletti, C. G., Brazzini, G., Piacentini, F., Passerini, S., Limongi, D., Ciotti, M., Palamara, A. T., Marfia, G. A., & Pietropaolo, V. (2021). Risk Assessment of Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis Patients during 1 Year of Ocrelizumab Treatment. Viruses, 13(9), 1684. https://doi.org/10.3390/v13091684







