DNA Helicase–Polymerase Coupling in Bacteriophage DNA Replication
Abstract
:1. Introduction
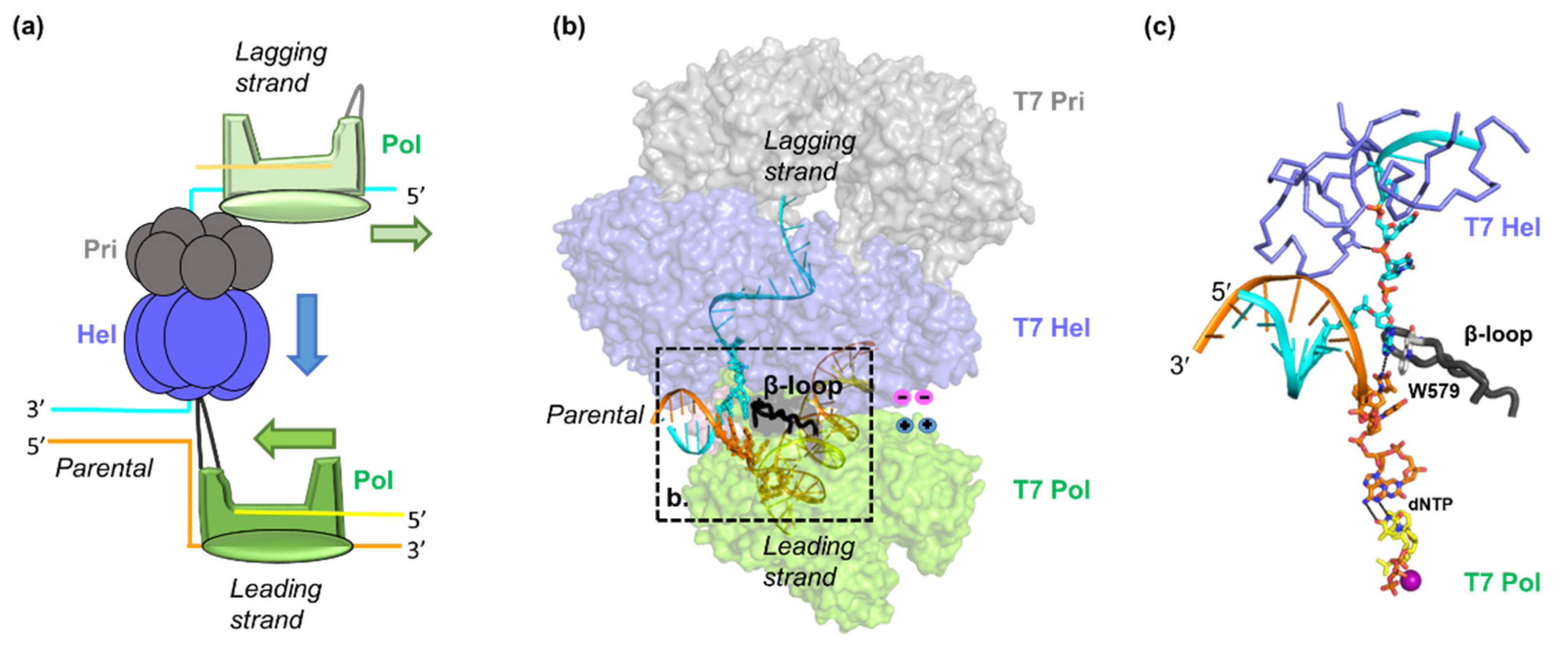
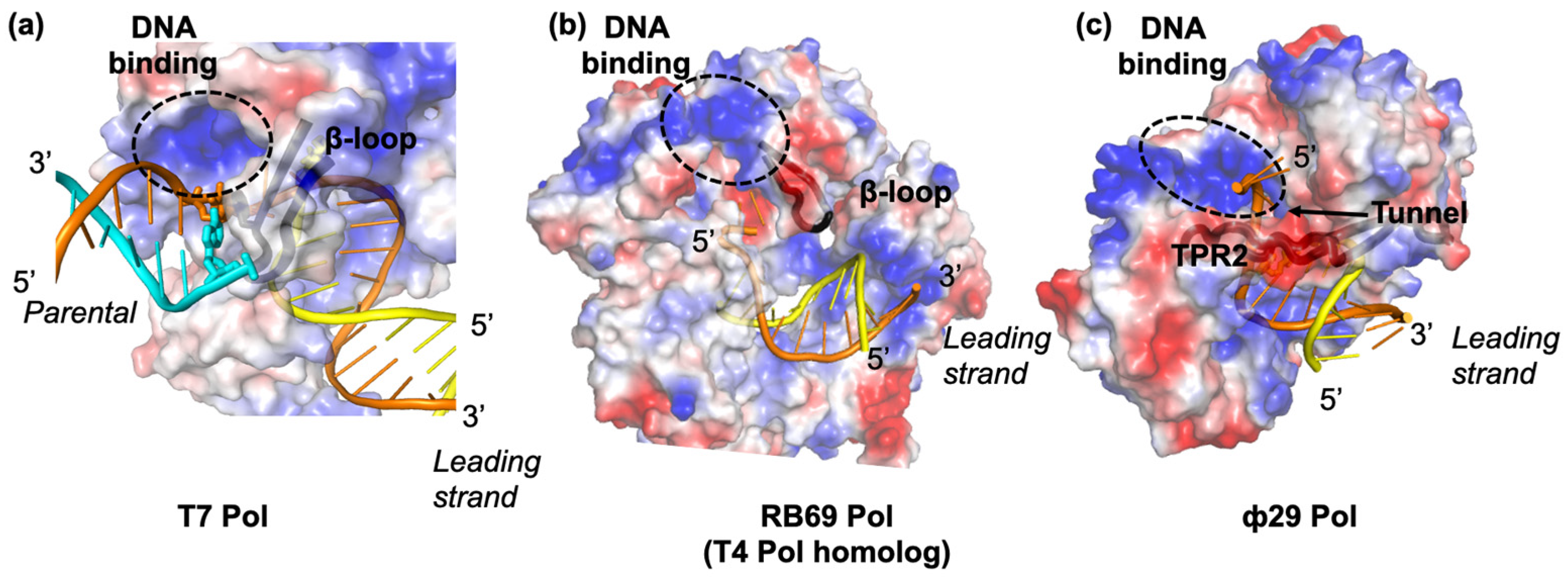
2. Hel–Pol Coupling in Bacteriophage T7 DNA Replication
3. Hel–Pol Coupling in Bacteriophage T4 DNA Replication
4. Bacteriophage Φ29 DNA Replication without a Helicase
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [Green Version]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Hershey, A.D.; Chase, M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 1952, 36, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.M.; Richardson, C.C. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009, 78, 205–243. [Google Scholar] [CrossRef] [Green Version]
- Weigel, C.; Seitz, H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006, 30, 321–381. [Google Scholar] [CrossRef] [PubMed]
- Benkovic, S.J.; Valentine, A.M.; Salinas, F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001, 70, 181–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, M.; Langston, L.; Stillman, B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Chaudhry, W.N.; Akhtar, M.N.; Andleeb, S.; Qadri, I. Bacteriophages and their implications on future biotechnology: A review. Virol. J. 2012, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.J.; Waksman, G. Structure and mechanism of DNA polymerases. Adv. Protein. Chem. 2005, 71, 401–440. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, S.M.; van Oijen, A.M. Timing, coordination, and rhythm: Acrobatics at the DNA replication fork. J. Biol. Chem. 2010, 285, 18979–18983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waga, S.; Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998, 67, 721–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, D. Proteomic Analyses of the Eukaryotic Replication Machinery. Methods. Enzymol. 2017, 591, 33–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dressler, D.; Wolfson, J.; Magazin, M. Initiation and reinitiation of DNA synthesis during replication of bacteriophage T7. Proc. Natl. Acad. Sci. USA 1972, 69, 998–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, T.J., Jr.; Thomas, C.A., Jr. An intermediate in the replication of bacteriophage T7 DNA molecules. J. Mol. Biol. 1969, 44, 459–475. [Google Scholar] [CrossRef]
- Doublie, S.; Tabor, S.; Long, A.M.; Richardson, C.C.; Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 1998, 391, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Hamdan, S.M.; Cook, T.E.; Richardson, C.C. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J. Biol. Chem. 2008, 283, 32077–32084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modrich, P.; Richardson, C.C. Bacteriophage T7 Deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J. Biol. Chem. 1975, 250, 5508–5514. [Google Scholar] [CrossRef]
- Huber, H.E.; Russel, M.; Model, P.; Richardson, C.C. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 1986, 261, 15006–15012. [Google Scholar] [CrossRef]
- Zhang, H.; Lee, S.J.; Kulczyk, A.W.; Zhu, B.; Richardson, C.C. Heterohexamer of 56- and 63-kDa Gene 4 Helicase-Primase of Bacteriophage T7 in DNA Replication. J. Biol. Chem. 2012, 287, 34273–34287. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Cui, Y.; Fox, T.; Lin, S.; Wang, H.; de Val, N.; Zhou, Z.H.; Yang, W. Structures and operating principles of the replisome. Science 2019, 363. [Google Scholar] [CrossRef]
- Pandey, M.; Syed, S.; Donmez, I.; Patel, G.; Ha, T.; Patel, S.S. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature 2009, 462, 940–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, A.J.; Richardson, C.C. Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein. Semin. Cell Dev. Biol. 2019, 86, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, D.; Pandey, M.; Patel, S.S. Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. Elife 2015, 4. [Google Scholar] [CrossRef]
- Pandey, M.; Patel, S.S. Helicase and polymerase move together close to the fork junction and copy DNA in one-nucleotide steps. Cell Rep. 2014, 6, 1129–1138. [Google Scholar] [CrossRef] [Green Version]
- Stano, N.M.; Jeong, Y.J.; Donmez, I.; Tummalapalli, P.; Levin, M.K.; Patel, S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 2005, 435, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Pandey, M.; Nandakumar, D.; Raney, K.D.; Yin, Y.W.; Patel, S.S. Excessive excision of correct nucleotides during DNA synthesis explained by replication hurdles. EMBO J. 2020, 39, e103367. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.S.; Bai, L.; Smith, B.Y.; Patel, S.S.; Wang, M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 2007, 129, 1299–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, S.; Pandey, M.; Patel, S.S.; Ha, T. Single-molecule fluorescence reveals the unwinding stepping mechanism of replicative helicase. Cell Rep. 2014, 6, 1037–1045. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Chastain, P.D., 2nd; Kusakabe, T.; Griffith, J.D.; Richardson, C.C. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell 1998, 1, 1001–1010. [Google Scholar] [CrossRef]
- Hamdan, S.M.; Loparo, J.J.; Takahashi, M.; Richardson, C.C.; van Oijen, A.M. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature 2009, 457, 336–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamdan, S.M.; Marintcheva, B.; Cook, T.; Lee, S.J.; Tabor, S.; Richardson, C.C. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. USA 2005, 102, 5096–5101. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lee, S.J.; Zhu, B.; Tran, N.Q.; Tabor, S.; Richardson, C.C. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 9372–9377. [Google Scholar] [CrossRef] [Green Version]
- Hamdan, S.M.; Johnson, D.E.; Tanner, N.A.; Lee, J.B.; Qimron, U.; Tabor, S.; van Oijen, A.M.; Richardson, C.C. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell 2007, 27, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Juarez-Quintero, V.; Peralta-Castro, A.; Benitez Cardoza, C.G.; Ellenberger, T.; Brieba, L.G. Structure of an open conformation of T7 DNA polymerase reveals novel structural features regulating primer-template stabilization at the polymerization active site. Biochem. J. 2021, 478, 2665–2679. [Google Scholar] [CrossRef] [PubMed]
- Crampton, D.J.; Mukherjee, S.; Richardson, C.C. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell 2006, 21, 165–174. [Google Scholar] [CrossRef]
- Sun, B.; Johnson, D.S.; Patel, G.; Smith, B.Y.; Pandey, M.; Patel, S.S.; Wang, M.D. ATP-induced helicase slippage reveals highly coordinated subunits. Nature 2011, 478, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Hogg, M.; Aller, P.; Konigsberg, W.; Wallace, S.S.; Doublie, S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2007, 282, 1432–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamtekar, S.; Berman, A.J.; Wang, J.; Lazaro, J.M.; de Vega, M.; Blanco, L.; Salas, M.; Steitz, T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage phi29. Mol. Cell 2004, 16, 609–618. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yang, W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 2006, 127, 1349–1360. [Google Scholar] [CrossRef] [Green Version]
- Pike, A.C.; Shrestha, B.; Popuri, V.; Burgess-Brown, N.; Muzzolini, L.; Costantini, S.; Vindigni, A.; Gileadi, O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc. Natl. Acad. Sci. USA 2009, 106, 1039–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosas, M.; Xi, X.G.; Bensimon, D.; Croquette, V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010, 38, 5518–5526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Pandey, M.; Inman, J.T.; Yang, Y.; Kashlev, M.; Patel, S.S.; Wang, M.D. T7 replisome directly overcomes DNA damage. Nat. Commun. 2015, 6, 10260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Singh, A.; Sultana, S.; Inman, J.T.; Patel, S.S.; Wang, M.D. Helicase promotes replication re-initiation from an RNA transcript. Nat. Commun. 2018, 9, 2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulczyk, A.W.; Moeller, A.; Meyer, P.; Sliz, P.; Richardson, C.C. Cryo-EM structure of the replisome reveals multiple interactions coordinating DNA synthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E1848–E1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallen, J.R.; Zhang, H.; Weis, C.; Cui, W.; Foster, B.M.; Ho, C.M.W.; Hammel, M.; Tainer, J.A.; Gross, M.L.; Ellenberger, T. Hybrid Methods Reveal Multiple Flexibly Linked DNA Polymerases within the Bacteriophage T7 Replisome. Structure 2017, 25, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [Green Version]
- Noble, E.; Spiering, M.M.; Benkovic, S.J. Coordinated DNA Replication by the Bacteriophage T4 Replisome. Viruses 2015, 7, 3186–3200. [Google Scholar] [CrossRef] [PubMed]
- Benkovic, S.J.; Spiering, M.M. Understanding DNA replication by the bacteriophage T4 replisome. J. Biol. Chem. 2017, 292, 18434–18442. [Google Scholar] [CrossRef] [PubMed]
- Mace, D.C.; Alberts, B.M. T4 DNA polymerase. Rates and processivity on single-stranded DNA templates. J. Mol. Biol. 1984, 177, 295–311. [Google Scholar] [CrossRef]
- Young, M.C.; Schultz, D.E.; Ring, D.; von Hippel, P.H. Kinetic parameters of the translocation of bacteriophage T4 gene 41 protein helicase on single-stranded DNA. J. Mol. Biol. 1994, 235, 1447–1458. [Google Scholar] [CrossRef]
- Jing, D.H.; Dong, F.; Latham, G.J.; von Hippel, P.H. Interactions of bacteriophage T4-coded primase (gp61) with the T4 replication helicase (gp41) and DNA in primosome formation. J. Biol. Chem. 1999, 274, 27287–27298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishmael, F.T.; Alley, S.C.; Benkovic, S.J. Assembly of the bacteriophage T4 helicase: Architecture and stoichiometry of the gp41-gp59 complex. J. Biol. Chem. 2002, 277, 20555–20562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosas, M.; Spiering, M.M.; Ding, F.; Croquette, V.; Benkovic, S.J. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012, 40, 6187–6198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosas, M.; Spiering, M.M.; Ding, F.; Bensimon, D.; Allemand, J.F.; Benkovic, S.J.; Croquette, V. Mechanism of strand displacement synthesis by DNA replicative polymerases. Nucleic Acids Res. 2012, 40, 6174–6186. [Google Scholar] [CrossRef]
- Lionnet, T.; Spiering, M.M.; Benkovic, S.J.; Croquette, V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 19790–19795. [Google Scholar] [CrossRef] [Green Version]
- Norcum, M.T.; Warrington, J.A.; Spiering, M.M.; Ishmael, F.T.; Trakselis, M.A.; Benkovic, S.J. Architecture of the bacteriophage T4 primosome: Electron microscopy studies of helicase (gp41) and primase (gp61). Proc. Natl. Acad. Sci. USA 2005, 102, 3623–3626. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sattar, A.K.; Wang, C.C.; Karam, J.D.; Konigsberg, W.H.; Steitz, T.A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 1997, 89, 1087–1099. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Konigsberg, W.H. RB69 DNA polymerase structure, kinetics, and fidelity. Biochemistry 2014, 53, 2752–2767. [Google Scholar] [CrossRef]
- Munn, M.M.; Alberts, B.M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J. Biol. Chem. 1991, 266, 20034–20044. [Google Scholar] [CrossRef]
- Delagoutte, E.; von Hippel, P.H. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry 2001, 40, 4459–4477. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Zhang, Z.; Zhuang, Z.; Yang, J.; Spiering, M.M.; Hammes, G.G.; Benkovic, S.J. Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): Unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome. Biochemistry 2005, 44, 7747–7756. [Google Scholar] [CrossRef]
- Martin, G.; Lazaro, J.M.; Mendez, E.; Salas, M. Characterization of the phage phi 29 protein p5 as a single-stranded DNA binding protein. Function in phi 29 DNA-protein p3 replication. Nucleic Acids Res. 1989, 17, 3663–3672. [Google Scholar] [CrossRef] [Green Version]
- Salas, M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991, 60, 39–71. [Google Scholar] [CrossRef]
- Pastrana, R.; Lazaro, J.M.; Blanco, L.; Garcia, J.A.; Mendez, E.; Salas, M. Overproduction and purification of protein P6 of Bacillus subtilis phage phi 29: Role in the initiation of DNA replication. Nucleic Acids Res. 1985, 13, 3083–3100. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.; Bernad, A.; Lazaro, J.M.; Martin, G.; Garmendia, C.; Salas, M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989, 264, 8935–8940. [Google Scholar] [CrossRef]
- Morin, J.A.; Cao, F.J.; Lazaro, J.M.; Arias-Gonzalez, J.R.; Valpuesta, J.M.; Carrascosa, J.L.; Salas, M.; Ibarra, B. Active DNA unwinding dynamics during processive DNA replication. Proc. Natl. Acad. Sci. USA 2012, 109, 8115–8120. [Google Scholar] [CrossRef] [Green Version]
- Morin, J.A.; Cao, F.J.; Lazaro, J.M.; Arias-Gonzalez, J.R.; Valpuesta, J.M.; Carrascosa, J.L.; Salas, M.; Ibarra, B. Mechano-chemical kinetics of DNA replication: Identification of the translocation step of a replicative DNA polymerase. Nucleic Acids Res. 2015, 43, 3643–3652. [Google Scholar] [CrossRef] [Green Version]
- Dufour, E.; Mendez, J.; Lazaro, J.M.; de Vega, M.; Blanco, L.; Salas, M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000, 304, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Spenkelink, L.M.; Spinks, R.R.; Jergic, S.; Lewis, J.S.; Dixon, N.E.; van Oijen, A.M. The E. coli helicase does not use ATP during replication. bioRxiv 2021. [Google Scholar] [CrossRef]
- Georgescu, R.; Yuan, Z.; Bai, L.; de Luna Almeida Santos, R.; Sun, J.; Zhang, D.; Yurieva, O.; Li, H.; O’Donnell, M.E. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. USA 2017, 114, E697–E706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berti, M.; Cortez, D.; Lopes, M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell. Biol. 2020, 21, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Lain, S.; Riechmann, J.L.; Garcia, J.A. RNA helicase: A novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acids Res. 1990, 18, 7003–7006. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, C.-Y.; Gao, Y. DNA Helicase–Polymerase Coupling in Bacteriophage DNA Replication. Viruses 2021, 13, 1739. https://doi.org/10.3390/v13091739
Lo C-Y, Gao Y. DNA Helicase–Polymerase Coupling in Bacteriophage DNA Replication. Viruses. 2021; 13(9):1739. https://doi.org/10.3390/v13091739
Chicago/Turabian StyleLo, Chen-Yu, and Yang Gao. 2021. "DNA Helicase–Polymerase Coupling in Bacteriophage DNA Replication" Viruses 13, no. 9: 1739. https://doi.org/10.3390/v13091739
APA StyleLo, C.-Y., & Gao, Y. (2021). DNA Helicase–Polymerase Coupling in Bacteriophage DNA Replication. Viruses, 13(9), 1739. https://doi.org/10.3390/v13091739





