Interplay between Host tRNAs and HIV-1: A Structural Perspective
Abstract
:1. Introduction
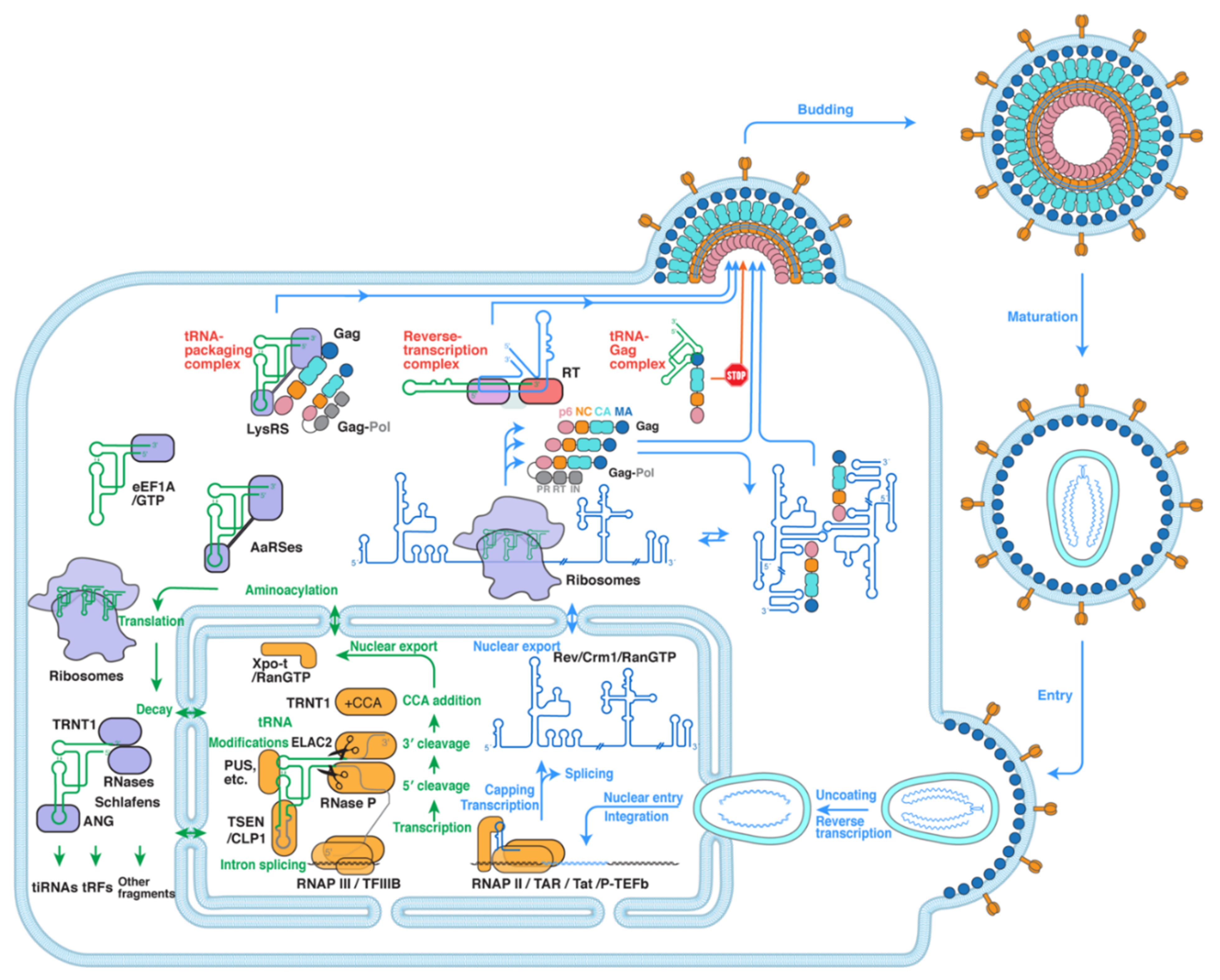
2. The Interwoven Paths of Host tRNAs and HIV-1
3. Exploitation of tRNA as Reverse Transcription Primers
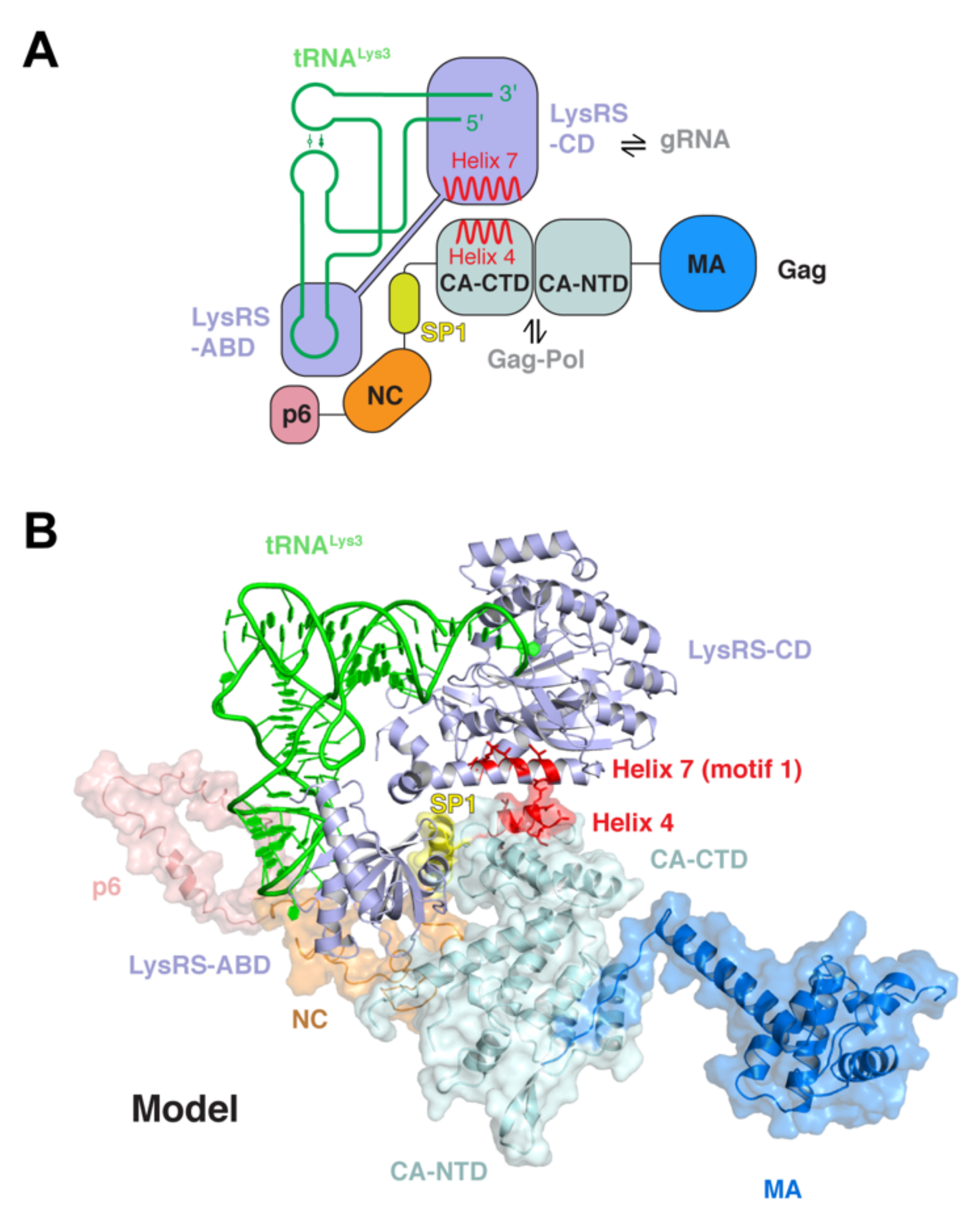
4. Hijacking and Packaging of tRNA Primers into HIV Virions
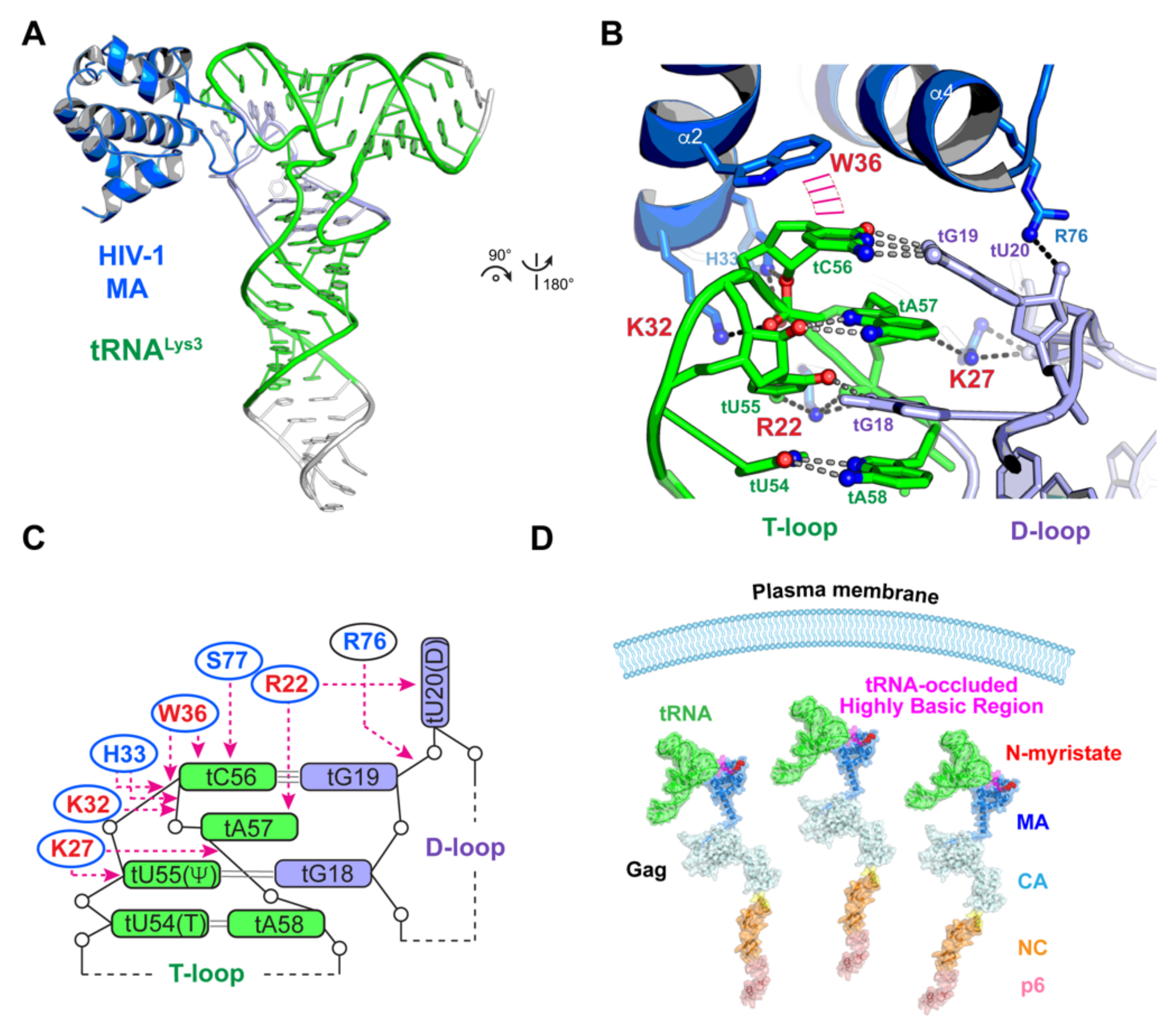
5. Viral Appropriation of Host tRNAs to Regulate Gag Localization
6. Role of tRNAs and Associated Proteins in Host Defense against HIV-1
7. Summary, Conclusions and Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korostelev, A.; Trakhanov, S.; Laurberg, M.; Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 2006, 126, 1065–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cate, J.H.; Yusupov, M.M.; Yusupova, G.Z.; Earnest, T.N.; Noller, H.F. X-ray crystal structures of 70S ribosome functional complexes. Science 1999, 285, 2095–2104. [Google Scholar] [CrossRef]
- Nissen, P.; Hansen, J.; Ban, N.; Moore, P.B.; Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 2000, 289, 920–930. [Google Scholar] [CrossRef] [Green Version]
- Steitz, T.A.; Moore, P.B. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 2003, 28, 411–418. [Google Scholar] [CrossRef]
- Selmer, M.; Dunham, C.M.; Murphy, F.V., IV; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 2002, 108, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Martinez, A.; Yamashita, S.; Tomita, K. Crystal structure of human cytoplasmic tRNAHis-specific 5’-monomethylphosphate capping enzyme. Nucleic Acids Res. 2020, 48, 1572–1582. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, T.; Miyauchi, K.; Sakaguchi, Y.; Yamashita, S.; Soma, A.; Tomita, K.; Suzuki, T. Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat. Chem. Biol. 2018, 14, 1010–1020. [Google Scholar] [CrossRef]
- Suzuki, T.; Yashiro, Y.; Kikuchi, I.; Ishigami, Y.; Saito, H.; Matsuzawa, I.; Okada, S.; Mito, M.; Iwasaki, S.; Ma, D.; et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020, 11, 4269. [Google Scholar] [CrossRef]
- Söll, D. Transfer RNA: An RNA for all seasons. In The RNA World; Gesteland, R.F., Atkins, J.F., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; pp. 157–184. [Google Scholar]
- Ibba, M.; Söll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000, 69, 617–650. [Google Scholar] [CrossRef] [PubMed]
- Ribas de Pouplana, L.; Schimmel, P. Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem. Sci. 2001, 26, 591–596. [Google Scholar] [CrossRef]
- Schimmel, P.; Beebe, K. Aminoacyl tRNA synthases: From the RNA world to the theater of proteins. In The RNA World, 3rd ed.; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 2006; pp. 227–255. [Google Scholar]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Lo, W.S.; Gardiner, E.; Xu, Z.; Lau, C.F.; Wang, F.; Zhou, J.J.; Mendlein, J.D.; Nangle, L.A.; Chiang, K.P.; Yang, X.L.; et al. Human tRNA synthetase catalytic nulls with diverse functions. Science 2014, 345, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Hamann, C.S.; Hou, Y.M. Probing a tRNA core that contributes to aminoacylation. J. Mol. Biol. 2000, 295, 777–789. [Google Scholar] [CrossRef]
- Jin, D.; Musier-Forsyth, K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J. Biol. Chem. 2019, 294, 5352–5364. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, A.E.; Pinkard, O.; Coller, J.M. tRNA metabolism and neurodevelopmental disorders. Annu Rev. Genom. Hum. Genet. 2019, 20, 359–387. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baer, M.; Nilsen, T.W.; Costigan, C.; Altman, S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990, 18, 97–103. [Google Scholar] [CrossRef]
- Mondragon, A. Structural studies of RNase P. Annu Rev. Biophys. 2013, 42, 537–557. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, S.; Rosch, S.; Marchfelder, A. Assigning a function to a conserved group of proteins: The tRNA 3’-processing enzymes. EMBO J. 2002, 21, 2769–2777. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.R.; Kessler, A.C.; Polycarpo, C.; Alfonzo, J.D. Cutting, dicing, healing and sealing: The molecular surgery of tRNA. Wiley Interdiscip. Rev. RNA 2015, 6, 337–349. [Google Scholar] [CrossRef] [Green Version]
- Yoo, C.J.; Wolin, S.L. The yeast La protein is required for the 3’ endonucleolytic cleavage that matures tRNA precursors. Cell 1997, 89, 393–402. [Google Scholar] [CrossRef] [Green Version]
- Lizano, E.; Scheibe, M.; Rammelt, C.; Betat, H.; Morl, M. A comparative analysis of CCA-adding enzymes from human and E. coli: Differences in CCA addition and tRNA 3’-end repair. Biochimie 2008, 90, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Weiner, A.M. Collaboration between CC- and A-adding enzymes to build and repair the 3’-terminal CCA of tRNA in Aquifex aeolicus. Science 2001, 294, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Ishitani, R.; Fukai, S.; Nureki, O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature 2006, 443, 956–960. [Google Scholar] [CrossRef]
- Paushkin, S.V.; Patel, M.; Furia, B.S.; Peltz, S.W.; Trotta, C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell 2004, 117, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Zhuang, Y.; Zhu, C.; Meng, H.; Lu, B.; Xie, B.; Peng, J.; Li, M.; Yi, C. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 2020, 16, 160–169. [Google Scholar] [CrossRef]
- Cook, A.G.; Fukuhara, N.; Jinek, M.; Conti, E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 2009, 461, 60–65. [Google Scholar] [CrossRef]
- Lipowsky, G.; Bischoff, F.R.; Izaurralde, E.; Kutay, U.; Schafer, S.; Gross, H.J.; Beier, H.; Gorlich, D. Coordination of tRNA nuclear export with processing of tRNA. RNA 1999, 5, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Guy, M.P.; Young, D.L.; Payea, M.J.; Zhang, X.; Kon, Y.; Dean, K.M.; Grayhack, E.J.; Mathews, D.H.; Fields, S.; Phizicky, E.M. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014, 28, 1721–1732. [Google Scholar] [CrossRef] [Green Version]
- Whipple, J.M.; Lane, E.A.; Chernyakov, I.; D’Silva, S.; Phizicky, E.M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 2011, 25, 1173–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhang, Y.; Zhou, T.; Chen, Q. tsRNAs: The swiss army knife for translational regulation. Trends Biochem. Sci. 2019, 44, 185–189. [Google Scholar] [CrossRef]
- Yue, T.; Zhan, X.; Zhang, D.; Jain, R.; Wang, K.W.; Choi, J.H.; Misawa, T.; Su, L.; Quan, J.; Hildebrand, S.; et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 2021, 372, 6543. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Sasikumar, A.N.; Perez, W.B.; Kinzy, T.G. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip. Rev. RNA 2012, 3, 543–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nature 2011, 469, 424–427. [Google Scholar] [CrossRef]
- Sabo, Y.; Walsh, D.; Barry, D.S.; Tinaztepe, S.; de Los Santos, K.; Goff, S.P.; Gundersen, G.G.; Naghavi, M.H. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 2013, 14, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Delaney, M.K.; Malikov, V.; Chai, Q.; Zhao, G.; Naghavi, M.H. Distinct functions of diaphanous-related formins regulate HIV-1 uncoating and transport. Proc. Natl. Acad. Sci. USA 2017, 114, E6932–E6941. [Google Scholar] [CrossRef] [Green Version]
- Dharan, A.; Opp, S.; Abdel-Rahim, O.; Keceli, S.K.; Imam, S.; Diaz-Griffero, F.; Campbell, E.M. Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc. Natl. Acad. Sci. USA 2017, 114, E10707–E10716. [Google Scholar] [CrossRef] [Green Version]
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493. [Google Scholar] [CrossRef]
- Zila, V.; Margiotta, E.; Turonova, B.; Muller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Borner, K.; Rada, J.; Muller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18. [Google Scholar] [CrossRef]
- Schulze-Gahmen, U.; Hurley, J.H. Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. USA 2018, 115, 12973–12978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef] [Green Version]
- Emery, A.; Swanstrom, R. HIV-1: To Splice or Not to Splice, That Is the Question. Viruses 2021, 13, 181. [Google Scholar] [CrossRef]
- Fang, X.; Wang, J.; O’Carroll, I.P.; Mitchell, M.; Zuo, X.; Wang, Y.; Yu, P.; Liu, Y.; Rausch, J.W.; Dyba, M.A.; et al. An unusual topological structure of the HIV-1 Rev response element. Cell 2013, 155, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Kharytonchyk, S.; Brown, J.D.; Stilger, K.; Yasin, S.; Iyer, A.S.; Collins, J.; Summers, M.F.; Telesnitsky, A. Influence of gag and RRE Sequences on HIV-1 RNA Packaging Signal Structure and Function. J. Mol. Biol. 2018, 430, 2066–2079. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010, 17, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Kharytonchyk, S.; Monti, S.; Smaldino, P.J.; Van, V.; Bolden, N.C.; Brown, J.D.; Russo, E.; Swanson, C.; Shuey, A.; Telesnitsky, A.; et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13378–13383. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.D.; Kharytonchyk, S.; Chaudry, I.; Iyer, A.S.; Carter, H.; Becker, G.; Desai, Y.; Glang, L.; Choi, S.H.; Singh, K.; et al. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 2020, 368, 413–417. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Kräusslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Saad, J.S. The interplay between HIV-1 gag binding to the plasma membrane and env incorporation. Viruses 2020, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K. Maturation of retroviruses. Curr. Opin. Virol. 2019, 36, 47–55. [Google Scholar] [CrossRef]
- Larsen, K.P.; Mathiharan, Y.K.; Kappel, K.; Coey, A.T.; Chen, D.H.; Barrero, D.; Madigan, L.; Puglisi, J.D.; Skiniotis, G.; Puglisi, E.V. Architecture of an HIV-1 reverse transcriptase initiation complex. Nature 2018, 557, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, L.; Jones, C.P.; Musier-Forsyth, K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010, 584, 359–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bou-Nader, C.; Muecksch, F.; Brown, J.B.; Gordon, J.M.; York, A.; Peng, C.; Ghirlando, R.; Summers, M.F.; Bieniasz, P.D.; Zhang, J. HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe 2021, 29, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.S.; Hughes, S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012, 2, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Workowski, K.A.; Bolan, G.A.; Centers for Disease. Prevention, sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep. 2015, 64, 1–137. [Google Scholar]
- Smith, A.J.; Cho, M.I.; Hammarskjold, M.L.; Rekosh, D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 1990, 64, 2743–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [Green Version]
- Ilina, T.V.; Slack, R.L.; Elder, J.H.; Sarafianos, S.G.; Parniak, M.A.; Ishima, R. Effect of tRNA on the Maturation of HIV-1 Reverse Transcriptase. J. Mol. Biol. 2018, 430, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- Slack, R.L.; Ilina, T.V.; Xi, Z.; Giacobbi, N.S.; Kawai, G.; Parniak, M.A.; Sarafianos, S.G.; Sluis Cremer, N.; Ishima, R. Conformational Changes in HIV-1 Reverse Transcriptase that Facilitate Its Maturation. Structure 2019, 27, 1581–1593.e3. [Google Scholar] [CrossRef] [PubMed]
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag-membrane interactions by specific RNAs. RNA 2017, 23, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Gremminger, T.; Singh, G.; Cheng, Y.; Li, J.; Qiu, L.; Ji, J.; Lange, M.J.; Zuo, X.; Chen, S.J.; et al. The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity. Nucleic Acids Res. 2021, 49, 5925–5942. [Google Scholar] [CrossRef]
- Telesnitsky, A.; Wolin, S.L. The Host RNAs in Retroviral Particles. Viruses 2016, 8, 235. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.B.; Yildiz, F.Z.; Lo, J.A.; Wang, B.; D’Souza, V.M. A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature 2014, 515, 591–595. [Google Scholar] [CrossRef] [Green Version]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014, 454, 362–370. [Google Scholar] [CrossRef] [Green Version]
- Bieniasz, P.; Telesnitsky, A. Multiple, switchable protein: RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 2018, 5, 165–183. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beerens, N.; Groot, F.; Berkhout, B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001, 276, 31247–31256. [Google Scholar] [CrossRef] [Green Version]
- Beerens, N.; Berkhout, B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 2002, 76, 2329–2339. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J. Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1600. [Google Scholar] [CrossRef]
- Zhang, J.; Ferré-D’Amaré, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 2013, 500, 363–366. [Google Scholar] [CrossRef]
- Li, S.; Su, Z.; Lehmann, J.; Stamatopoulou, V.; Giarimoglou, N.; Henderson, F.E.; Fan, L.; Pintilie, G.D.; Zhang, K.; Chen, M.; et al. Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat. Struct. Mol. Biol. 2019, 26, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferré-D’Amaré, A.R. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol. Cell 2014, 55, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Suddala, K.C.; Zhang, J. High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove. Nat. Struct. Mol. Biol. 2019, 26, 1114–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckwahl, M.J.; Arnion, H.; Kharytonchyk, S.; Zang, T.; Bieniasz, P.D.; Telesnitsky, A.; Wolin, S.L. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 2016, 22, 1228–1238. [Google Scholar] [CrossRef]
- Itano, M.S.; Arnion, H.; Wolin, S.L.; Simon, S.M. Recruitment of 7SL RNA to assembling HIV-1 virus-like particles. Traffic 2018, 19, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tian, C.; Zhang, W.; Luo, K.; Sarkis, P.T.; Yu, L.; Liu, B.; Yu, Y.; Yu, X.F. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007, 81, 13112–13124. [Google Scholar] [CrossRef] [Green Version]
- Kleiman, L.; Cen, S. The tRNALys packaging complex in HIV-1. Int. J. Biochem. Cell Biol. 2004, 36, 1776–1786. [Google Scholar] [CrossRef]
- Khorchid, A.; Javanbakht, H.; Wise, S.; Halwani, R.; Parniak, M.A.; Wainberg, M.A.; Kleiman, L. Sequences within Pr160gag-pol affecting the selective packaging of primer tRNA(Lys3) into HIV-1. J. Mol. Biol. 2000, 299, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kovaleski, B.J.; Kennedy, R.; Khorchid, A.; Kleiman, L.; Matsuo, H.; Musier-Forsyth, K. Critical role of helix 4 of HIV-1 capsid C-terminal domain in interactions with human lysyl-tRNA synthetase. J. Biol. Chem. 2007, 282, 32274–32279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javanbakht, H.; Halwani, R.; Cen, S.; Saadatmand, J.; Musier-Forsyth, K.; Gottlinger, H.; Kleiman, L. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 2003, 278, 27644–27651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Shapiro, R.; Morris, G.M.; Yang, X.L.; Schimmel, P. Packaging HIV virion components through dynamic equilibria of a human tRNA synthetase. J. Phys. Chem. B 2010, 114, 16273–16279. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.P.; Cantara, W.A.; Olson, E.D.; Musier-Forsyth, K. Small-angle X-ray scattering-derived structure of the HIV-1 5’ UTR reveals 3D tRNA mimicry. Proc. Natl. Acad. Sci. USA 2014, 111, 3395–3400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.P.; Saadatmand, J.; Kleiman, L.; Musier-Forsyth, K. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 2013, 19, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Comandur, R.; Jones, C.P.; Tsang, P.; Musier-Forsyth, K. Anticodon-like binding of the HIV-1 tRNA-like element to human lysyl-tRNA synthetase. RNA 2016, 22, 1828–1835. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef] [Green Version]
- Mercredi, P.Y.; Bucca, N.; Loeliger, B.; Gaines, C.R.; Mehta, M.; Bhargava, P.; Tedbury, P.R.; Charlier, L.; Floquet, N.; Muriaux, D.; et al. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016, 428, 1637–1655. [Google Scholar] [CrossRef] [Green Version]
- Gaines, C.R.; Tkacik, E.; Rivera-Oven, A.; Somani, P.; Achimovich, A.; Alabi, T.; Zhu, A.; Getachew, N.; Yang, A.L.; McDonough, M.; et al. HIV-1 matrix protein interactions with tRNA: Implications for membrane targeting. J. Mol. Biol. 2018, 430, 2113–2127. [Google Scholar] [CrossRef]
- Göttlinger, H.G.; Sodroski, J.G.; Haseltine, W.A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 1989, 86, 5781–5785. [Google Scholar] [CrossRef] [Green Version]
- Bryant, M.; Ratner, L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 1990, 87, 523–527. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Parent, L.J.; Wills, J.W.; Resh, M.D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 1994, 68, 2556–2569. [Google Scholar] [CrossRef] [Green Version]
- Hill, C.P.; Worthylake, D.; Bancroft, D.P.; Christensen, A.M.; Sundquist, W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 1996, 93, 3099–3104. [Google Scholar] [CrossRef] [Green Version]
- Tang, C.; Ndassa, Y.; Summers, M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002, 9, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Massiah, M.A.; Starich, M.R.; Paschall, C.; Summers, M.F.; Christensen, A.M.; Sundquist, W.I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 1994, 244, 198–223. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, R.A.; Kamynina, E.; Vogt, V.M. Effect of multimerization on membrane association of Rous sarcoma virus and HIV-1 matrix domain proteins. J. Virol. 2013, 87, 13598–13608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Caballero, D.; Hatziioannou, T.; Martin-Serrano, J.; Bieniasz, P.D. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on gag precursor-membrane interactions. J. Virol. 2004, 78, 9560–9563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monje-Galvan, V.; Voth, G.A. Binding mechanism of the matrix domain of HIV-1 gag on lipid membranes. Elife 2020, 9, e58621. [Google Scholar] [CrossRef]
- Van der Kuyl, A.C.; Berkhout, B. The biased nucleotide composition of the HIV genome: A constant factor in a highly variable virus. Retrovirology 2012, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Keating, C.P.; Hill, M.K.; Hawkes, D.J.; Smyth, R.P.; Isel, C.; Le, S.Y.; Palmenberg, A.C.; Marshall, J.A.; Marquet, R.; Nabel, G.J.; et al. The A-rich RNA sequences of HIV-1 pol are important for the synthesis of viral cDNA. Nucleic Acids Res. 2009, 37, 945–956. [Google Scholar] [CrossRef]
- Jakobsen, M.R.; Mogensen, T.H.; Paludan, S.R. Caught in translation: Innate restriction of HIV mRNA translation by a schlafen family protein. Cell Res. 2013, 23, 320–322. [Google Scholar] [CrossRef] [Green Version]
- Holmes, M.; Zhang, F.; Bieniasz, P.D. Single-cell and single-cycle analysis of HIV-1 replication. PLoS Pathog. 2015, 11, e1004961. [Google Scholar] [CrossRef] [Green Version]
- Nunes, A.; Ribeiro, D.R.; Marques, M.; Santos, M.A.S.; Ribeiro, D.; Soares, A.R. Emerging Roles of tRNAs in RNA Virus Infections. Trends Biochem. Sci. 2020, 45, 794–805. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.Y.; Deng, X.Y.; Li, Y.S.; Ma, X.C.; Feng, J.X.; Yu, B.; Chen, Y.; Luo, Y.L.; Wang, X.; Chen, M.L.; et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018, 9, 1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterlin, B.M.; Liu, P.; Wang, X.; Cary, D.; Shao, W.; Leoz, M.; Hong, T.; Pan, T.; Fujinaga, K. Hili inhibits HIV replication in activated T cells. J. Virol. 2017, 91, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Qiu, H.; Garcia-Barrio, M.; Anderson, J.; Hinnebusch, A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 2000, 6, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Lageix, S.; Zhang, J.; Rothenburg, S.; Hinnebusch, A.G. Interaction between the tRNA-binding and C-terminal domains of Yeast Gcn2 regulates kinase activity in vivo. PLoS Genet. 2015, 11, e1004991. [Google Scholar] [CrossRef] [Green Version]
- Del Pino, J.; Jimenez, J.L.; Ventoso, I.; Castello, A.; Munoz-Fernandez, M.A.; de Haro, C.; Berlanga, J.J. GCN2 has inhibitory effect on human immunodeficiency virus-1 protein synthesis and is cleaved upon viral infection. PLoS ONE 2012, 7, e47272. [Google Scholar] [CrossRef] [PubMed]
- Jaspart, A.; Calmels, C.; Cosnefroy, O.; Bellecave, P.; Pinson, P.; Claverol, S.; Guyonnet-Duperat, V.; Dartigues, B.; Benleulmi, M.S.; Mauro, E.; et al. GCN2 phosphorylates HIV-1 integrase and decreases HIV-1 replication by limiting viral integration. Sci. Rep. 2017, 7, 2283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokladal, L.; Stumpe, M.; Pillet, B.; Hu, Z.; Garcia Osuna, G.M.; Kressler, D.; Dengjel, J.; De Virgilio, C. Global phosphoproteomics pinpoints uncharted Gcn2-mediated mechanisms of translational control. Mol. Cell 2021, 81, 1879–1889.e6. [Google Scholar] [CrossRef]
- Hood, I.V.; Gordon, J.M.; Bou-Nader, C.; Henderson, F.E.; Bahmanjah, S.; Zhang, J. Crystal structure of an adenovirus virus-associated RNA. Nat. Commun. 2019, 10, 2871. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Gordon, J.M.; Henderson, F.E.; Zhang, J. The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators. RNA 2019, 25, 539–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clerzius, G.; Gelinas, J.F.; Gatignol, A. Multiple levels of PKR inhibition during HIV-1 replication. Rev. Med. Virol. 2011, 21, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Gunnery, S.; Rice, A.P.; Robertson, H.D.; Mathews, M.B. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 1990, 87, 8687–8691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edery, I.; Petryshyn, R.; Sonenberg, N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: A novel translational control mechanism. Cell 1989, 56, 303–312. [Google Scholar] [CrossRef]
- Ofir-Birin, Y.; Fang, P.; Bennett, S.P.; Zhang, H.M.; Wang, J.; Rachmin, I.; Shapiro, R.; Song, J.; Dagan, A.; Pozo, J.; et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 2013, 49, 30–42. [Google Scholar] [CrossRef] [Green Version]
- Halwani, R.; Cen, S.; Javanbakht, H.; Saadatmand, J.; Kim, S.; Shiba, K.; Kleiman, L. Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 2004, 78, 7553–7564. [Google Scholar] [CrossRef] [Green Version]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Osswein, M.; Glass, B.; Mucksch, F.; Muller, R.; Schultz, C.; Muller, B.; Krausslich, H.G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Bukrinsky, M.I.; Haggerty, S.; Dempsey, M.P.; Sharova, N.; Adzhubel, A.; Spitz, L.; Lewis, P.; Goldfarb, D.; Emerman, M.; Stevenson, M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 1993, 365, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 gag forms ribonucleoprotein complexes with unspliced viral RNA at transcription sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef]
- Yannay-Cohen, N.; Carmi-Levy, I.; Kay, G.; Yang, C.M.; Han, J.M.; Kemeny, D.M.; Kim, S.; Nechushtan, H.; Razin, E. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 2009, 34, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Duchon, A.A.; St Gelais, C.; Titkemeier, N.; Hatterschide, J.; Wu, L.; Musier-Forsyth, K. HIV-1 Exploits a Dynamic Multi-aminoacyl-tRNA Synthetase Complex To Enhance Viral Replication. J. Virol. 2017, 91, 21. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J. Interplay between Host tRNAs and HIV-1: A Structural Perspective. Viruses 2021, 13, 1819. https://doi.org/10.3390/v13091819
Zhang J. Interplay between Host tRNAs and HIV-1: A Structural Perspective. Viruses. 2021; 13(9):1819. https://doi.org/10.3390/v13091819
Chicago/Turabian StyleZhang, Jinwei. 2021. "Interplay between Host tRNAs and HIV-1: A Structural Perspective" Viruses 13, no. 9: 1819. https://doi.org/10.3390/v13091819
APA StyleZhang, J. (2021). Interplay between Host tRNAs and HIV-1: A Structural Perspective. Viruses, 13(9), 1819. https://doi.org/10.3390/v13091819




