The Isolated in Utero Environment Is Conducive to the Emergence of RNA and DNA Virus Variants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of RNA and DNA Virus Stocks
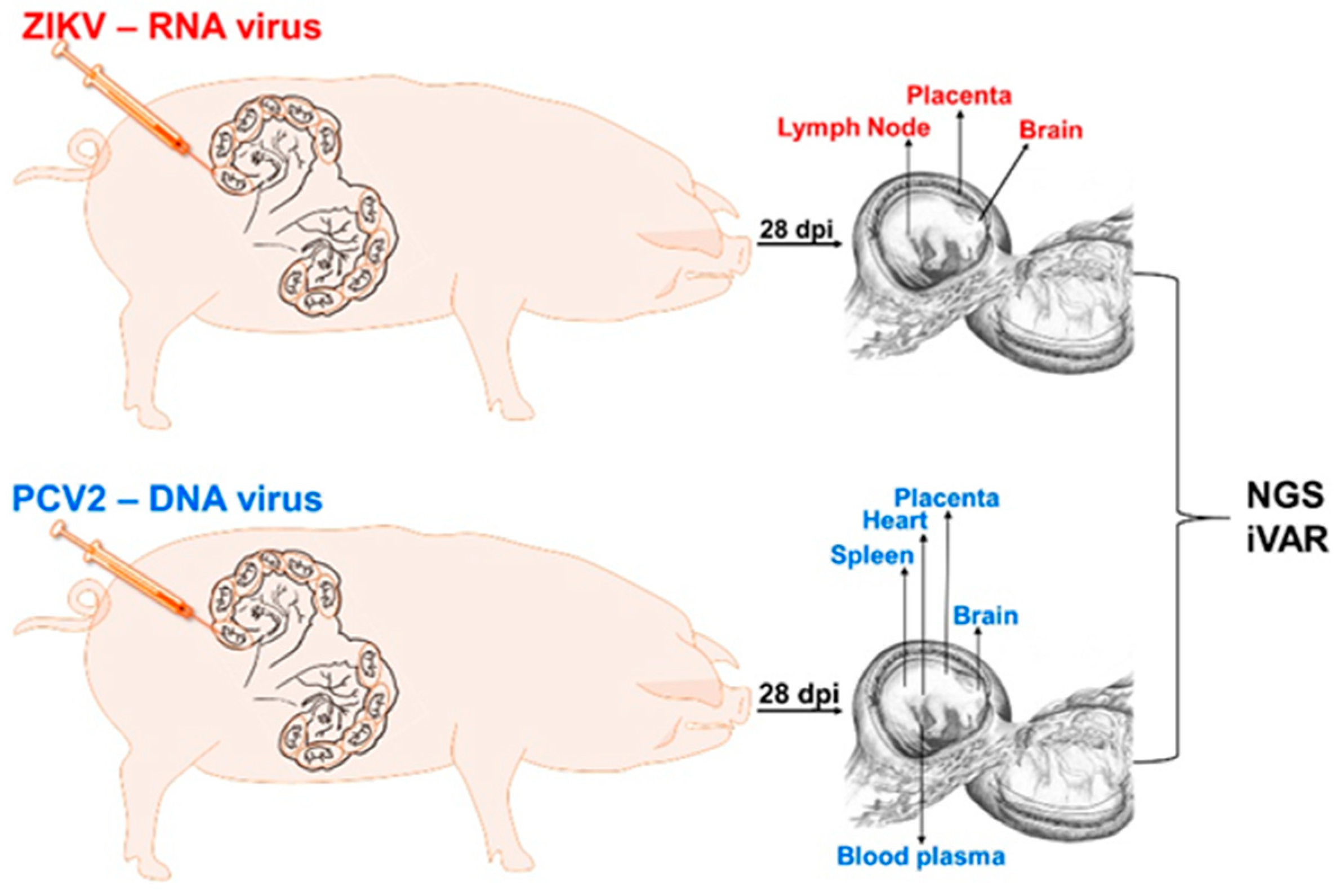
2.2. Animal Experiments
2.3. Quantification of ZIKV Loads and Virus-Specific Antibodies
2.4. Quantification of PCV2 Loads and Virus-Specific Antibodies
2.5. Next-Generation Sequencing
2.6. Illumina Data Processing and Variant Calling Using iVar
2.7. Statistical Analysis
3. Results
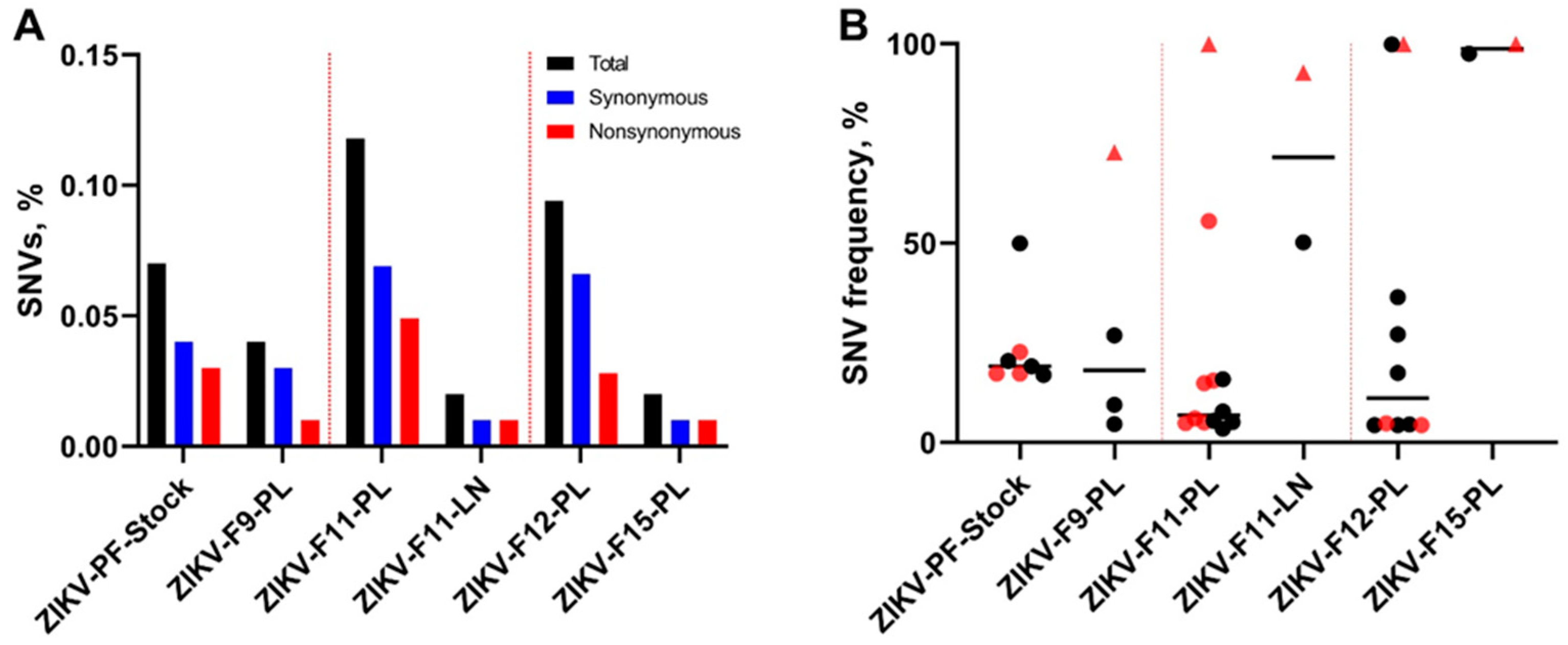
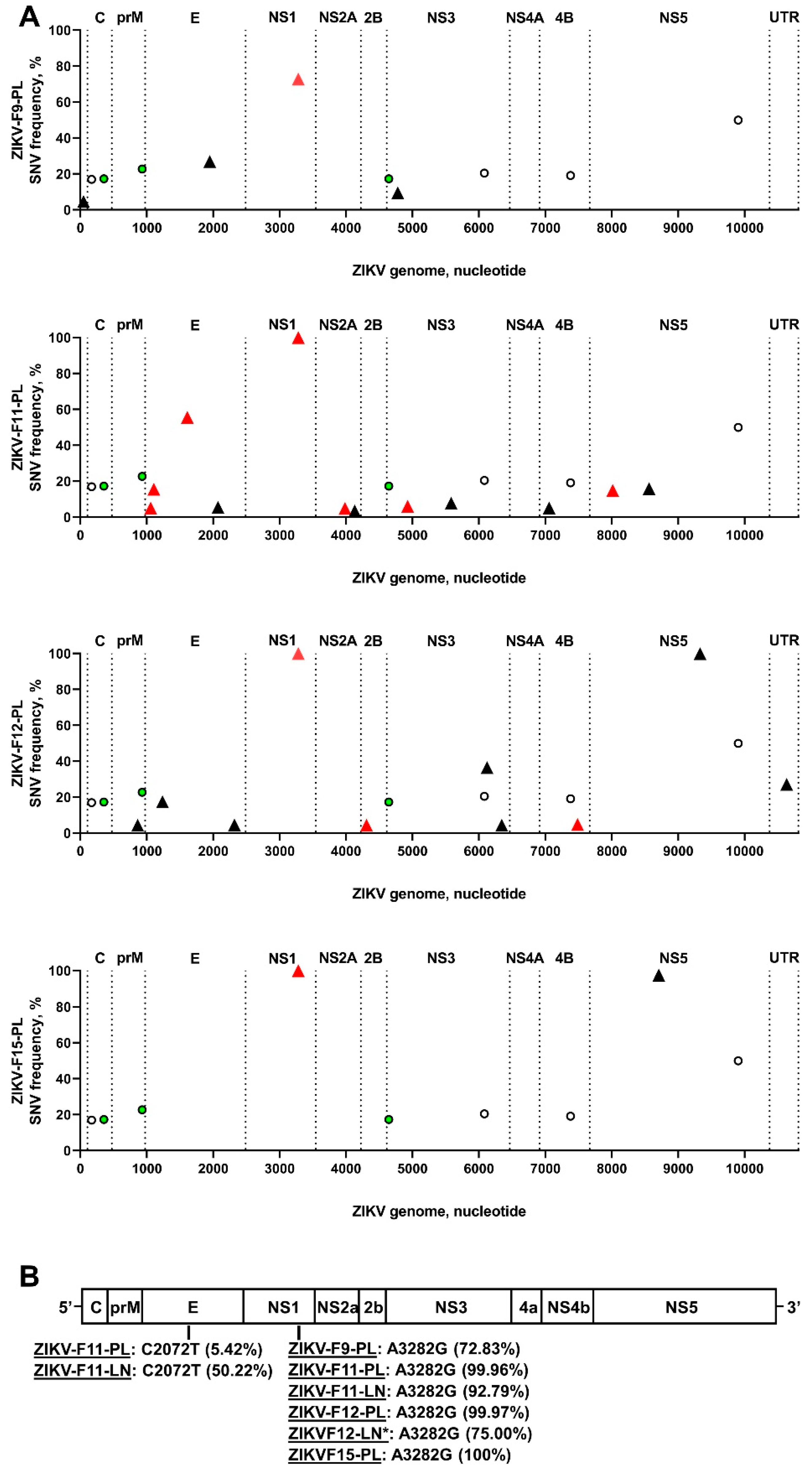
3.1. Zika Virus Variants Emerge during In Utero Infection
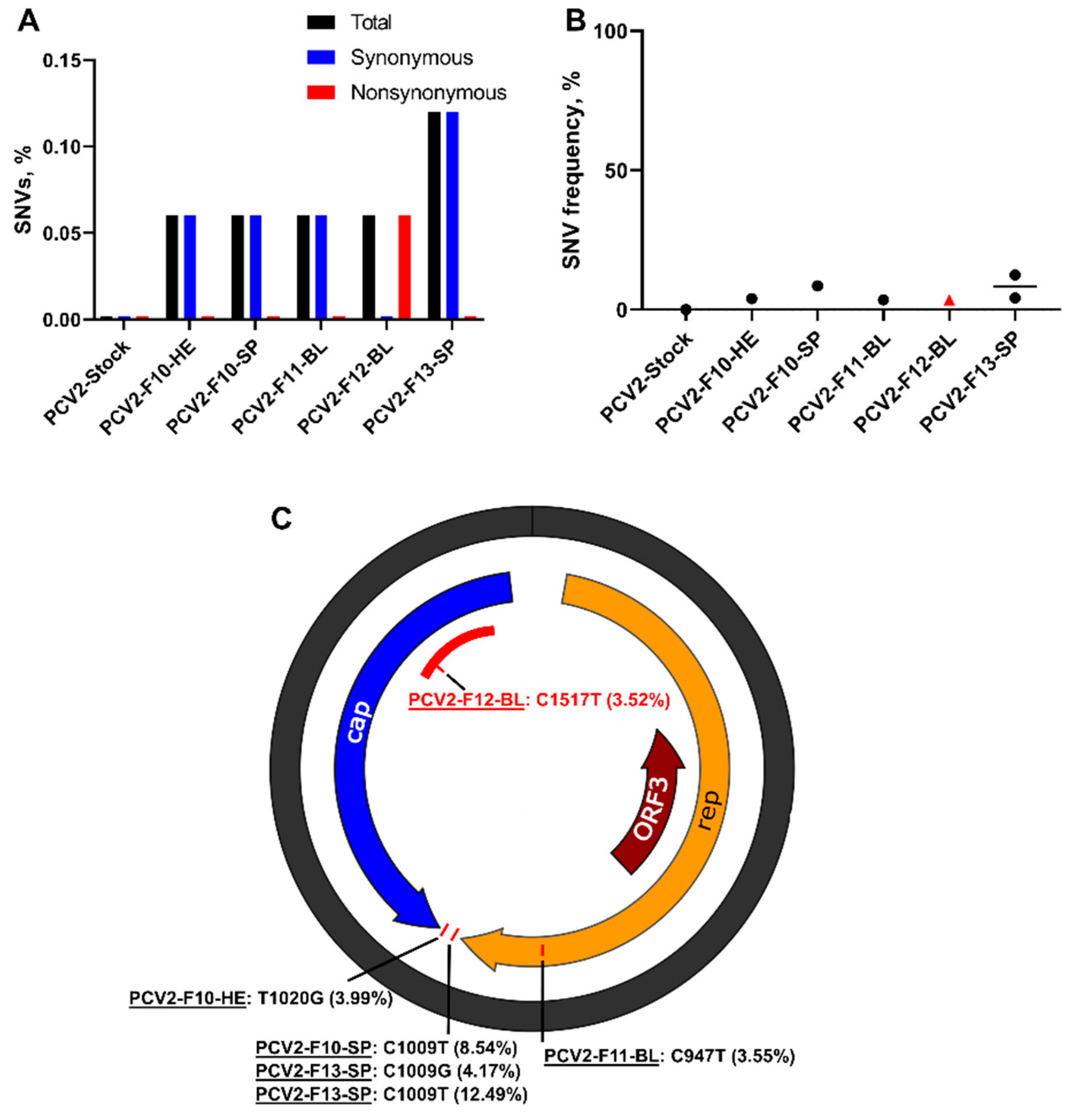
3.2. Porcine Circovirus 2 Variants Emerge during In Utero Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engels, G.; Hierweger, A.M.; Hoffmann, J.; Thieme, R.; Thiele, S.; Bertram, S.; Dreier, C.; Resa-Infante, P.; Jacobsen, H.; Thiele, K.; et al. Pregnancy-Related Immune Adaptation Promotes the Emergence of Highly Virulent H1N1 Influenza Virus Strains in Allogenically Pregnant Mice. Cell Host Microbe 2017, 21, 321–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dow, N.; Chernick, A.; Orsel, K.; Van Marle, G.; Van Der Meer, F. Genetic variability of bovine viral diarrhea virus and evidence for a possible genetic bottleneck during vertical transmission in persistently infected cattle. PLoS ONE 2015, 10, e131972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladinig, A.; Wilkinson, J.; Ashley, C.; Detmer, S.E.; Lunney, J.K.; Plastow, G.; Harding, J.C.S. Variation in fetal outcome, viral load and ORF5 sequence mutations in a large scale study of phenotypic responses to late gestation exposure to type 2 porcine reproductive and respiratory syndrome virus. PLoS ONE 2014, 9, e96104. [Google Scholar] [CrossRef] [PubMed]
- Rowland, R.R.R. The interaction between PRRSV and the late gestation pig fetus. Virus Res. 2010, 154, 114–122. [Google Scholar] [CrossRef]
- Menu, E.; M’Bopi Kéou, F.X.; Lagaye, S.; Pissard, S.; Mauclère, P.; Scarlatti, G.; Martin, J.; Goossens, M.; Chaouat, G.; Barré-Sinoussi, F. Selection of maternal human immunodeficiency virus type 1 variants in human placenta. J. Infect. Dis. 1999, 179, 44–51. [Google Scholar] [CrossRef]
- Trus, I.; Udenze, D.; Berube, N.; Wheler, C.; Martel, M.-J.; Gerdts, V.; Karniychuk, U. CpG-recoding in Zika virus genome causes host-age-dependent attenuation of infection with protection against lethal heterologous challenge in mice. Front. Immunol. 2019, 10, 3077. [Google Scholar] [CrossRef] [Green Version]
- Zakhartchouk, A.; Zhou, Y.; Tikoo, S.K. A recombinant E1-deleted porcine adenovirus-3 as an expression vector. Virology 2003, 313, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Steverink, P.J.G.M.; Van der Vorst, T.J.K.; Pesch, S.; Ohlinger, V.F.; Berndsen, F.W.; Hunneman, W.; Peperkamp, N.H.M.T.; Van Oirschot, J.T.; de Jong, M.F.; Schippers, R.; et al. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands. Vet. Q. 2000, 22, 167–172. [Google Scholar] [CrossRef]
- Udenze, D.; Trus, I.; Berube, N.; Gerdts, V.; Karniychuk, U. The African strain of Zika virus causes more severe in utero infection than Asian strain in a porcine fetal transmission model. Emerg. Microbes Infect. 2019, 8, 1098–1107. [Google Scholar] [CrossRef] [Green Version]
- Trus, I.; Darbellay, J.; Huang, Y.; Gilmour, M.; Safronetz, D.; Gerdts, V.; Karniychuk, U. Persistent Zika virus infection in porcine conceptuses is associated with elevated in utero cortisol levels. Virulence 2018, 9, 1338–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darbellay, J.; Lai, K.; Babiuk, S.; Berhane, Y.; Ambagala, A.; Wheler, C.; Wilson, D.; Walker, S.; Potter, A.; Gilmour, M.; et al. Neonatal pigs are susceptible to experimental Zika virus infection. Emerg. Microbes Infect. 2017, 6, e6. [Google Scholar] [CrossRef] [Green Version]
- Trus, I.; Udenze, D.; Cox, B.; Berube, N.; Nordquist, R.E.; van der Staay, F.J.; Huang, Y.; Kobinger, G.; Safronetz, D.; Gerdts, V.; et al. Subclinical in utero Zika virus infection is associated with interferon alpha sequelae and sex-specific molecular brain pathology in asymptomatic porcine offspring. PLOS Pathog. 2019, 15, e1008038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trus, I.; Walker, S.; Fuchs, M.; Udenze, D.; Gerdts, V.; Karniychuk, U. A Porcine Model of Zika Virus Infection to Profile the in Utero Interferon Alpha Response. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2142, pp. 181–195. [Google Scholar]
- Darbellay, J.; Cox, B.; Lai, K.; Delgado-Ortega, M.; Wheler, C.; Wilson, D.; Walker, S.; Starrak, G.; Hockley, D.; Huang, Y.; et al. Zika Virus Causes Persistent Infection in Porcine Conceptuses and may Impair Health in Offspring. EBioMedicine. 2017, 25, 73–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Del Pozo Sacristán, R.; Van Renne, N.; Huang, L.; Decaluwe, R.; Michiels, A.; Rodriguez, A.L.; Rodríguez, M.J.; Durán, M.G.; Declerk, I.; et al. Anti-porcine circovirus type 2 (PCV2) antibody placental barrier leakage from sow to fetus: Impact on the diagnosis of intra-uterine PCV2 infection. Virol. Sin. 2014, 29, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Karniychuk, U.U.; Huang, L.; Geldhof, M.; Vanhee, M.; Lefebvre, D.J.; Meerts, P.; Ducatelle, R.; Doorsselaere, J.V.; Nauwynck, H.J. Unusual outcome of in utero infection and subsequent postnatal super-infection with different PCV2b strains. Virol. Sin. 2014, 29, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Lefebvre, D.J.; Van Doorsselaere, J.; Atanasova, K.; Barbé, F.; Geldhof, M.; Karniychuk, U.U.; Nauwynck, H.J. Pathologic and virologic findings in mid-gestational porcine foetuses after experimental inoculation with PCV2a or PCV2b. Vet. Microbiol. 2010, 145, 62–68. [Google Scholar] [CrossRef]
- Saha, D.; Lefebvre, D.J.; Ducatelle, R.; Doorsselaere, J.V.; Nauwynck, H.J. Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses. BMC Vet. Res. 2011, 7, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.Y.; Liu, S.Q.; Deng, C.L.; Zhang, Q.Y.; Zhang, B. Detection of Zika virus by SYBR green one-step real-time RT-PCR. J. Virol. Methods 2016, 236, 93–97. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Nadeau, H.; Maxted, M.; Peregrine, J.; Reuter, D.; Norris, A.; Edwards, R.; Hyatt, K.; Singleton, K.; Papin, J.F.; et al. Maternal Zika Virus (ZIKV) Infection following Vaginal Inoculation with ZIKV-Infected Semen in Timed-Pregnant Olive Baboons. J. Virol. 2020, 94, e00058-20. [Google Scholar] [CrossRef] [Green Version]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Gröber, C.; Blank, M.; Händler, K.; Beyer, M.; Schultze, J.L.; Mayer, G. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, M.G.; Russ, C.; Costello, M.; Hollinger, A.; Lennon, N.J.; Hegarty, R.; Nusbaum, C.; Jaffe, D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trible, B.R.; Kerrigan, M.; Crossland, N.; Potter, M.; Faaberg, K.; Hesse, R.; Rowland, R.R.R. Antibody recognition of porcine circovirus type 2 capsid protein epitopes after vaccination, infection, and disease. Clin. Vaccine Immunol. 2011, 18, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Trible, B.R.; Rowland, R.R.R. Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis. Virus Res. 2012, 164, 68–77. [Google Scholar] [CrossRef]
- Rosenberg, A.Z.; Yu, W.; Hill, D.A.; Reyes, C.A.; Schwartz, D.A. Placental Pathology of Zika Virus: Viral Infection of the Placenta Induces Villous Stromal Macrophage (Hofbauer Cell) Proliferation and Hyperplasia. Arch. Pathol. Lab. Med. 2017, 141, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Lind, N.M.; Moustgaard, A.; Jelsing, J.; Vajta, G.; Cumming, P.; Hansen, A.K. The use of pigs in neuroscience: Modeling brain disorders. Neurosci. Biobehav. Rev. 2007, 31, 728–751. [Google Scholar] [CrossRef]
- Johnson, K.E.E.; Noval, M.G.; Rangel, M.V.; De Jesus, E.; Geber, A.; Schuster, S.; Cadwell, K.; Ghedin, E.; Stapleford, K.A. Mapping the evolutionary landscape of Zika virus infection in immunocompromised mice. Virus Evol. 2020, 6, veaa092. [Google Scholar] [CrossRef]
- Caires-Júnior, L.C.; Goulart, E.; Melo, U.S.; Araujo, B.H.S.; Alvizi, L.; Soares-Schanoski, A.; de Oliveira, D.F.; Kobayashi, G.S.; Griesi-Oliveira, K.; Musso, C.M.; et al. Discordant congenital Zika syndrome twins show differential in vitro viral susceptibility of neural progenitor cells. Nat. Commun. 2018, 9, 475. [Google Scholar] [CrossRef] [Green Version]
- Ramos, R.C.; Brainer-Lima, A.M.; Cordeiro, M.T.; van der Linden, H., Jr.; da Caudas Neto, S.S.; Ventura, L.O.; Rolim Filho, E.L.; Cruz, D.D.C.S.; Florêncio, T.L.T.; van der Linden, V.; et al. Discordant clinical outcomes of congenital Zika virus infection in twin pregnancies. Arq. Neuropsiquiatr. 2017, 75, 381–386. [Google Scholar] [CrossRef]
- Nunez-Avellaneda, D.; Cetina-Trejo, R.C.; Zamudio-Moreno, E.; Baak-Baak, C.; Cigarroa-Toledo, N.; Reyes-Solis, G.; Ortega-Pacheco, A.; Suzán, G.; Tandugu, C.; García-Rejón, J.E.; et al. Evidence of Zika Virus Infection in Pigs and Mosquitoes, Mexico. Emerg. Infect. Dis. 2021, 27, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Šterzl, J.; Rejnek, J.; Trávníček, J. Impermeability of pig placenta for antibodies. Folia Microbiol. 1966, 11, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Correa-Fiz, F.; Franzo, G.; Llorens, A.; Huerta, E.; Sibila, M.; Kekarainen, T.; Segalés, J. Porcine circovirus 2 (PCV2) population study in experimentally infected pigs developing PCV2-systemic disease or a subclinical infection. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.Y.S.; Krishnamurthy, S.R.; Pesavento, P.A.; McDermott, D.H.; et al. Discovery of several thousand highly diverse circular DNA viruses. Elife 2020, 9, e51971. [Google Scholar] [CrossRef] [PubMed]
- Ribbens, S.; Dewulf, J.; Koenen, F.; Laevens, H.; De Kruif, A. Transmission of classical swine fever. A review. Vet. Q. 2004, 26, 146–155. [Google Scholar] [CrossRef] [Green Version]
- Burns, K.F. Congenital Japanese B Encephalitis Infection of Swine. Exp. Biol. Med. 1950, 75, 621–625. [Google Scholar] [CrossRef] [Green Version]
- Desingu, P.A.; Ray, P.K.; Patel, B.H.M.; Singh, R.; Singh, R.K.; Saikumar, G. Pathogenic and Genotypic Characterization of a Japanese Encephalitis Virus Isolate Associated with Reproductive Failure in an Indian Pig Herd. PLoS ONE 2016, 11, e147611. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Udenze, D.; Trus, I.; Munyanduki, H.; Berube, N.; Karniychuk, U. The Isolated in Utero Environment Is Conducive to the Emergence of RNA and DNA Virus Variants. Viruses 2021, 13, 1827. https://doi.org/10.3390/v13091827
Udenze D, Trus I, Munyanduki H, Berube N, Karniychuk U. The Isolated in Utero Environment Is Conducive to the Emergence of RNA and DNA Virus Variants. Viruses. 2021; 13(9):1827. https://doi.org/10.3390/v13091827
Chicago/Turabian StyleUdenze, Daniel, Ivan Trus, Henry Munyanduki, Nathalie Berube, and Uladzimir Karniychuk. 2021. "The Isolated in Utero Environment Is Conducive to the Emergence of RNA and DNA Virus Variants" Viruses 13, no. 9: 1827. https://doi.org/10.3390/v13091827
APA StyleUdenze, D., Trus, I., Munyanduki, H., Berube, N., & Karniychuk, U. (2021). The Isolated in Utero Environment Is Conducive to the Emergence of RNA and DNA Virus Variants. Viruses, 13(9), 1827. https://doi.org/10.3390/v13091827





