The MAPK/ERK Pathway and the Role of DUSP1 in JCPyV Infection of Primary Astrocytes
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. siRNA Treatment
2.3. JCPyV Infection
2.4. Indirect Immunofluorescence Staining for Quantitation of JCPyV Infection
2.5. ICW Assay and LI-COR Quantification
2.6. Relative Quantification of DUSP1 Transcript Levels by qPCR
2.7. Preparation of Samples for RNA-Seq and RNA-Seq Analysis
2.8. STRING Interaction Database, GO Enrichment Analysis, and PANTHER Pathway Analysis
2.9. Statistical Analysis and Graphing in RStudio
3. Results
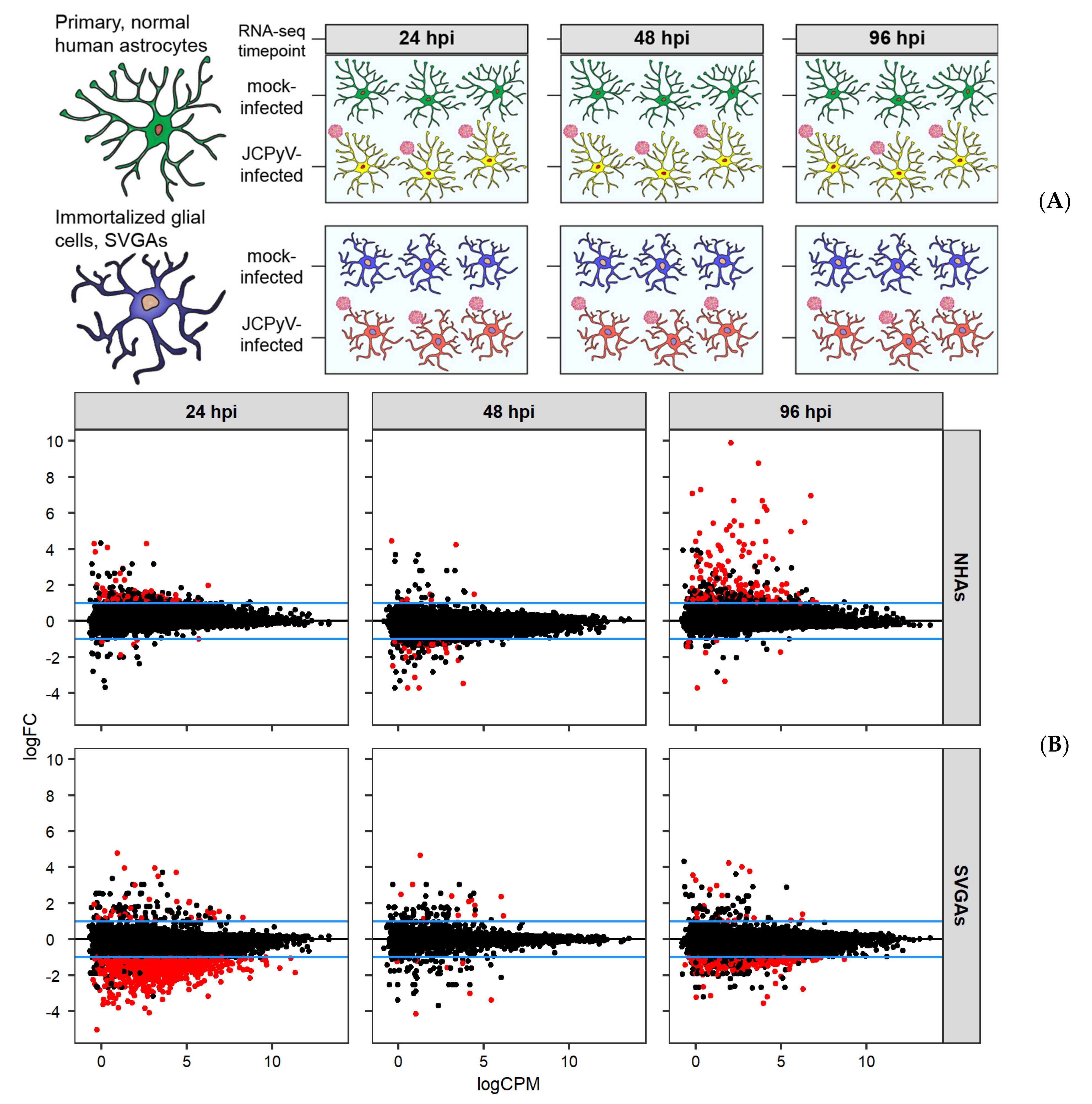
3.1. RNA-Seq Reveals Unique Differential Gene Expression in JCPyV-Infected Primary Astrocytes
3.2. The MAPK/ERK Pathway Is Differentially Expressed in NHAs during JCPyV Infection
3.3. JCPyV Requires ERK1/2 for Successful Infection in NHAs
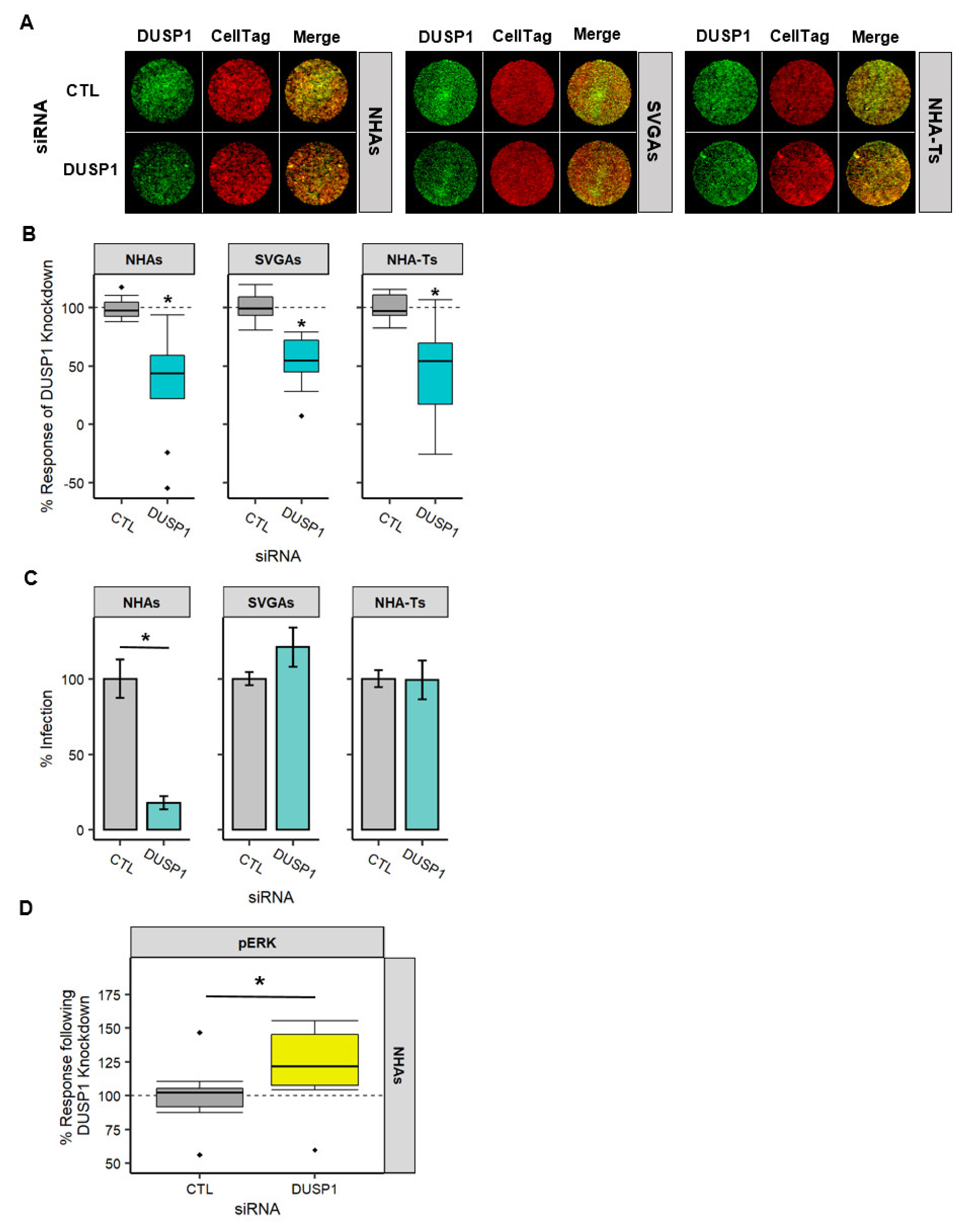
3.4. DUSP1 Transcript Decreases during JCPyV Infection in NHAs and Is Essential in Regulating the MAPK/ERK Pathway Compared to Immortalized Cells
3.5. DUSP1 Is Required for JCPyV Infection in NHAs
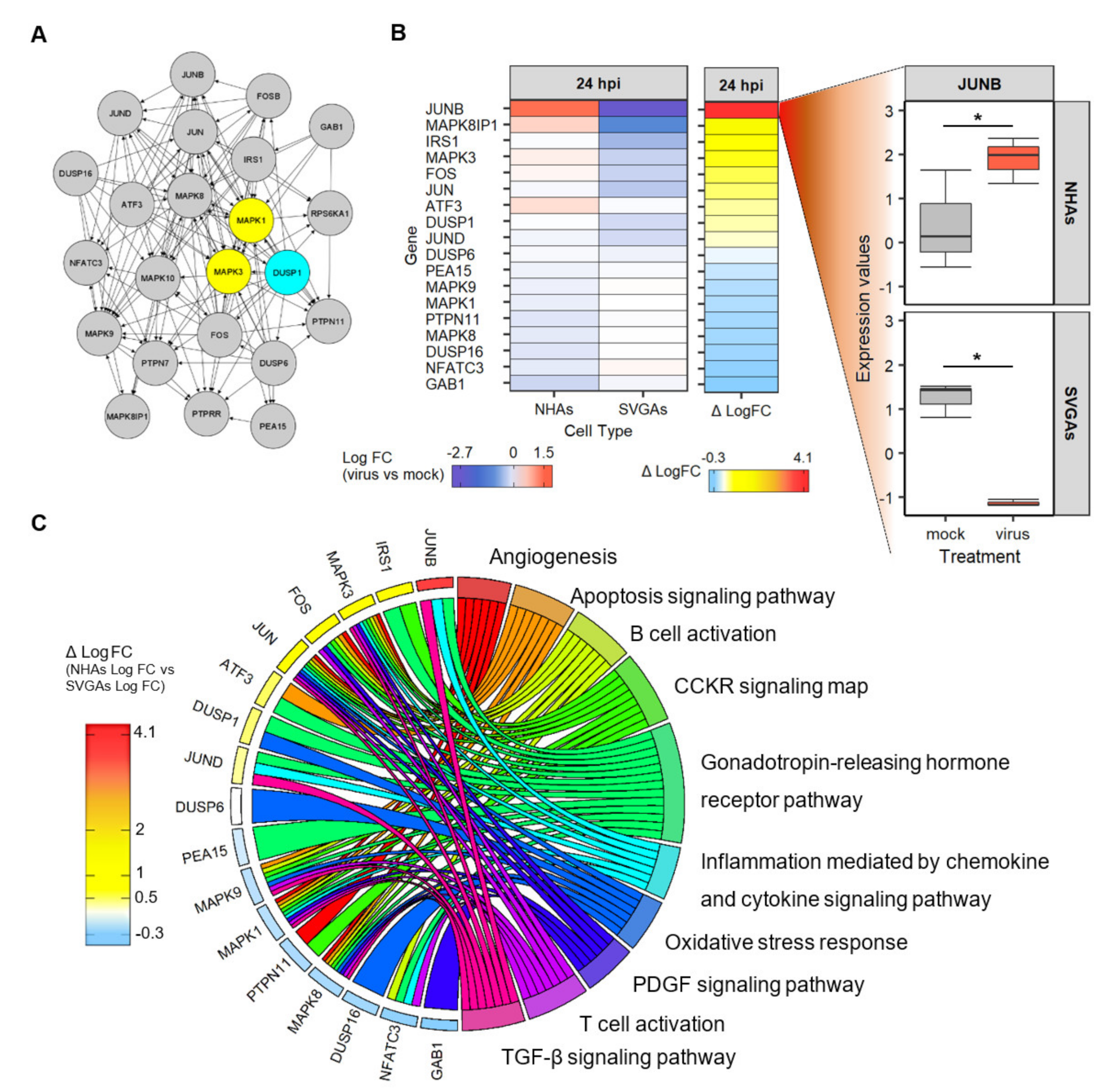
3.6. The Network of Genes Related to the Interactions of DUSP1 and ERK1/2 Are Involved in the Pathways of the Immune Response, Cell Survival, and Apoptosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirsch, H.H.; Kardas, P.; Kranz, D.; Leboeuf, C. The Human JC Polyomavirus (JCPyV): Virological Background and Clinical Implications. APMIS 2013, 121, 685–727. [Google Scholar] [CrossRef] [PubMed]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of Papova-like Virus from Human Brain with Progressive Multifocal Leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef]
- Silverman, L.; Rubinstein, L.J. Electron Microscopic Observations on a Case of Progressive Multifocal Leukoencephalopathy. Acta Neuropathol. 1965, 5, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Zurhein, G.; Chou, S.M. Particles Resembling Papova Viruses in Human Cerebral Demyelinating Disease. Science 1965, 148, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Windrem, M.S.; Zou, L.; Chandler-Militello, D.; Schanz, S.J.; Auvergne, R.M.; Betstadt, S.J.; Harrington, A.R.; Johnson, M.; Kazarov, A.; et al. Human Glial Chimeric Mice Reveal Astrocytic Dependence of JC Virus Infection. J. Clin. Investig. 2014, 124, 5323–5336. [Google Scholar] [CrossRef]
- Kean, J.M.; Rao, S.; Wang, M.; Garcea, R.L. Seroepidemiology of Human Polyomaviruses. PLoS Pathog. 2009, 5, e1000363. [Google Scholar] [CrossRef]
- Egli, A.; Infanti, L.; Dumoulin, A.; Buser, A.; Samaridis, J.; Stebler, C.; Gosert, R.; Hirsch, H.H. Prevalence of Polyomavirus BK and JC Infection and Replication in 400 Healthy Blood Donors. J. Infect. Dis. 2009, 199, 837–846. [Google Scholar] [CrossRef]
- Monaco, M.C.; Atwood, W.J.; Gravell, M.; Tornatore, C.S.; Major, E.O. JC Virus Infection of Hematopoietic Progenitor Cells, Primary B Lymphocytes, and Tonsillar Stromal Cells: Implications for Viral Latency. J. Virol 1996, 70, 7004–7012. [Google Scholar] [CrossRef]
- Dubois, V.; Dutronc, H.; Lafon, M.E.; Poinsot, V.; Pellegrin, J.L.; Ragnaud, J.M.; Ferrer, A.M.; Fleury, H.J. Latency and Reactivation of JC Virus in Peripheral Blood of Human Immunodeficiency Virus Type 1-Infected Patients. J. Clin. Microbiol. 1997, 35, 2288–2292. [Google Scholar] [CrossRef]
- Chapagain, M.L.; Nerurkar, V.R. Human Polyomavirus JC (JCV) Infection of Human B Lymphocytes: A Possible Mechanism for JCV Transmigration across the Blood-Brain Barrier. J. Infect. Dis. 2010, 202, 184–191. [Google Scholar] [CrossRef]
- Ferrante, P.; Caldarelli-Stefano, R.; Omodeo-Zorini, E.; Vago, L.; Boldorini, R.; Costanzi, G. PCR Detection of JC Virus DNA in Brain Tissue from Patients with and without Progressive Multifocal Leukoencephalopathy. J. Med. Virol. 1995, 47, 219–225. [Google Scholar] [CrossRef]
- Gorelik, L.; Reid, C.; Testa, M.; Brickelmaier, M.; Bossolasco, S.; Pazzi, A.; Bestetti, A.; Carmillo, P.; Wilson, E.; McAuliffe, M.; et al. Progressive Multifocal Leukoencephalopathy (PML) Development Is Associated with Mutations in JC Virus Capsid Protein VP1 That Change Its Receptor Specificity. J. Infect. Dis. 2011, 204, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Elzi, L.; Mueller, N.J.; Garzoni, C.; Cavassini, M.; Fux, C.A.; Vernazza, P.; Bernasconi, E.; Battegay, M.; Hirsch, H.H.; et al. Incidence and Outcome of Progressive Multifocal Leukoencephalopathy over 20 Years of the Swiss HIV Cohort Study. Clin. Infect. Dis. 2009, 48, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Carson, K.R.; Evens, A.M.; Richey, E.A.; Habermann, T.M.; Focosi, D.; Seymour, J.F.; Laubach, J.; Bawn, S.D.; Gordon, L.I.; Winter, J.N.; et al. Progressive Multifocal Leukoencephalopathy after Rituximab Therapy in HIV-Negative Patients: A Report of 57 Cases from the Research on Adverse Drug Events and Reports Project. Blood 2009, 113, 4834–4840. [Google Scholar] [CrossRef] [PubMed]
- Bloomgren, G.; Richman, S.; Hotermans, C.; Subramanyam, M.; Goelz, S.; Natarajan, A.; Lee, S.; Plavina, T.; Scanlon, J.V.; Sandrock, A.; et al. Risk of Natalizumab-Associated Progressive Multifocal Leukoencephalopathy. N. Engl. J. Med. 2012, 366, 1870–1880. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, D.; Patera, A.C.; Nyberg, F.; Gerber, M.; Liu, M. Leukeoncephalopathy Consortium Progressive Multifocal Leukoencephalopathy: Current Treatment Options and Future Perspectives. Ther. Adv. Neurol. Disord. 2015, 8, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.L.; Koralnik, I.J.; Rumbaugh, J.A.; Burger, P.C.; King-Rennie, A.; McArthur, J.C. Progressive Multifocal Leukoencephalopathy in a Patient without Immunodeficiency. Neurology 2011, 77, 297–299. [Google Scholar] [CrossRef]
- Vermersch, P.; Kappos, L.; Gold, R.; Foley, J.F.; Olsson, T.; Cadavid, D.; Bozic, C.; Richman, S. Clinical Outcomes of Natalizumab-Associated Progressive Multifocal Leukoencephalopathy (Podcast). Neurology 2011, 76, 1697–1704. [Google Scholar] [CrossRef]
- Prosperini, L.; de Rossi, N.; Scarpazza, C.; Moiola, L.; Cosottini, M.; Gerevini, S.; Capra, R. Italian PML Study Group Natalizumab-Related Progressive Multifocal Leukoencephalopathy in Multiple Sclerosis: Findings from an Italian Independent Registry. PLoS ONE 2016, 11, e0168376. [Google Scholar] [CrossRef]
- Seo, G.J.; Fink, L.H.L.; O’Hara, B.; Atwood, W.J.; Sullivan, C.S. Evolutionarily Conserved Function of a Viral MicroRNA. J. Virol. 2008, 82, 9823–9828. [Google Scholar] [CrossRef]
- Agostini, S.; Mancuso, R.; Costa, A.S.; Guerini, F.R.; Clerici, M. COS-7 Cells Are a Cellular Model to Monitor Polyomavirus JC MiR-J1-5p Expression. Mol. Biol. Rep. 2020, 47, 9201–9205. [Google Scholar] [CrossRef]
- Takahashi, K.; Sato, Y.; Sekizuka, T.; Kuroda, M.; Suzuki, T.; Hasegawa, H.; Katano, H. High Expression of JC Polyomavirus-Encoded MicroRNAs in Progressive Multifocal Leukoencephalopathy Tissues and Its Repressive Role in Virus Replication. PLoS Pathog. 2020, 16, e1008523. [Google Scholar] [CrossRef]
- Walker, D.L.; Padgett, B.L.; ZuRhein, G.M.; Albert, A.E.; Marsh, R.F. Human Papovavirus (JC): Induction of Brain Tumors in Hamsters. Science 1973, 181, 674–676. [Google Scholar] [CrossRef]
- Rhein, G.M.Z.; Varakis, J.N. Perinatal Induction of Medulloblastomas in Syrian Golden Hamsters by a Human Polyoma Virus (JC). Natl. Cancer Inst. Monogr. 1979, 51, 205–208. [Google Scholar]
- London, W.T.; Houff, S.A.; Madden, D.L.; Fuccillo, D.A.; Gravell, M.; Wallen, W.C.; Palmer, A.E.; Sever, J.L.; Padgett, B.L.; Walker, D.L.; et al. Brain Tumors in Owl Monkeys Inoculated with a Human Polyomavirus (JC Virus). Science 1978, 201, 1246–1249. [Google Scholar] [CrossRef] [PubMed]
- Houff, S.A.; London, W.T.; Rhein, G.M.Z.; Padgett, B.L.; Walker, D.L.; Sever, J.L. New World Primates as a Model of Viral-Induced Astrocytomas. Prog. Clin. Biol. Res. 1983, 105, 223–226. [Google Scholar] [PubMed]
- London, W.T.; Houff, S.A.; McKeever, P.E.; Wallen, W.C.; Sever, J.L.; Padgett, B.L.; Walker, D.L. Viral-Induced Astrocytomas in Squirrel Monkeys. Prog. Clin. Biol. Res. 1983, 105, 227–237. [Google Scholar]
- Major, E.O.; Vacante, D.A.; Traub, R.G.; London, W.T.; Sever, J.L. Owl Monkey Astrocytoma Cells in Culture Spontaneously Produce Infectious JC Virus Which Demonstrates Altered Biological Properties. J. Virol. 1987, 61, 1435–1441. [Google Scholar] [CrossRef]
- Major, E.O.; Miller, A.E.; Mourrain, P.; Traub, R.G.; de Widt, E.; Sever, J. Establishment of a Line of Human Fetal Glial Cells That Supports JC Virus Multiplication. Proc. Natl. Acad. Sci. USA 1985, 82, 1257–1261. [Google Scholar] [CrossRef]
- Lynch, K.J.; Frisque, R.J. Factors Contributing to the Restricted DNA Replicating Activity of JC Virus. Virology 1991, 180, 306–317. [Google Scholar] [CrossRef]
- Sock, E.; Wegner, M.; Fortunato, E.A.; Grummt, F. Large T-Antigen and Sequences within the Regulatory Region of JC Virus Both Contribute to the Features of JC Virus DNA Replication. Virology 1993, 197, 537–548. [Google Scholar] [CrossRef]
- Wilczek, M.P.; DuShane, J.K.; Armstrong, F.J.; Maginnis, M.S. JC Polyomavirus Infection Reveals Delayed Progression of the Infectious Cycle in Normal Human Astrocytes. J. Virol. 2019. [Google Scholar] [CrossRef]
- Erickson, K.D.; Garcea, R.L. Viral Replication Centers and the DNA Damage Response in JC Virus-Infected Cells. Virology 2019, 528, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Ferenczy, M.W.; Johnson, K.R.; Marshall, L.J.; Monaco, M.; Major, E.O. Differentiation of Human Fetal Multipotential Neural Progenitor Cells to Astrocytes Reveals Susceptibility Factors for JC Virus. J. Virol. 2013, 87, 6221–6231. [Google Scholar] [CrossRef] [PubMed]
- Mázló, M.; Tariska, I. Are Astrocytes Infected in Progressive Multifocal Leukoencephalopathy (PML)? Acta Neuropathol. 1982, 56, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.D.; Gordon, J.; Assimakopoulou, M.; Enam, S.; Geddes, J.F.; Varakis, J.N.; Katsetos, C.D.; Croul, S.; Khalili, K. Detection of JC Virus DNA Sequences and Expression of the Viral Regulatory Protein T-Antigen in Tumors of the Central Nervous System. Cancer Res. 2001, 61, 4287–4293. [Google Scholar] [PubMed]
- Ahuja, D.; Sáenz-Robles, M.T.; Pipas, J.M. SV40 Large T Antigen Targets Multiple Cellular Pathways to Elicit Cellular Transformation. Oncogene 2005, 24, 7729–7745. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Otte, J.; Enam, S.; Valle, L.D.; Khalili, K.; Gordon, J. JC Virus-Induced Changes in Cellular Gene Expression in Primary Human Astrocytes. J. Virol. 2003, 77, 10638–10644. [Google Scholar] [CrossRef]
- Assetta, B.; Cecco, M.D.; O’Hara, B.; Atwood, W.J. JC Polyomavirus Infection of Primary Human Renal Epithelial Cells Is Controlled by a Type I IFN-Induced Response. mBio 2016, 7, e00903-16. [Google Scholar] [CrossRef]
- DuShane, J.K.; Wilczek, M.P.; Mayberry, C.L.; Maginnis, M.S. ERK Is a Critical Regulator of JC Polyomavirus Infection. J. Virol. 2018, 92, e01529-17. [Google Scholar] [CrossRef]
- Querbes, W.; Benmerah, A.; Tosoni, D.; Fiore, P.P.D.; Atwood, W.J. A JC Virus-Induced Signal Is Required for Infection of Glial Cells by a Clathrin- and Eps15-Dependent Pathway. J. Virol. 2003, 78, 250–256. [Google Scholar] [CrossRef]
- Shaul, Y.D.; Seger, R. The MEK/ERK Cascade: From Signaling Specificity to Diverse Functions. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2007, 1773, 1213–1226. [Google Scholar] [CrossRef]
- DuShane, J.K.; Mayberry, C.L.; Wilczek, M.P.; Nichols, S.L.; Maginnis, M.S. JCPyV-Induced MAPK Signaling Activates Transcription Factors during Infection. Int. J. Mol. Sci. 2019, 20, 4779. [Google Scholar] [CrossRef]
- Pleschka, S. RNA Viruses and the Mitogenic Raf/MEK/ERK Signal Transduction Cascade. Biol. Chem. 2008, 389, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Panteva, M.; Korkaya, H.; Jameel, S. Hepatitis Viruses and the MAPK Pathway: Is This a Survival Strategy? Virus Res. 2003, 92, 131–140. [Google Scholar] [CrossRef]
- Bonjardim, C.A. Viral Exploitation of the MEK/ERK Pathway—A Tale of Vaccinia Virus and Other Viruses. Virology 2017, 507, 267–275. [Google Scholar] [CrossRef]
- Pérez-Sen, R.; Queipo, M.J.; Gil-Redondo, J.C.; Ortega, F.; Gómez-Villafuertes, R.; Miras-Portugal, M.T.; Delicado, E.G. Dual-Specificity Phosphatase Regulation in Neurons and Glial Cells. Int. J. Mol. Sci. 2019, 20, 1999. [Google Scholar] [CrossRef]
- Dickinson, R.J.; Keyse, S.M. Diverse Physiological Functions for Dual-Specificity MAP Kinase Phosphatases. J. Cell Sci. 2006, 119, 4607–4615. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Li, S.; Westermarck, J. Phosphatase-mediated Crosstalk between MAPK Signaling Pathways in the Regulation of Cell Survival. FASEB J. 2008, 22, 954–965. [Google Scholar] [CrossRef]
- Patterson, K.I.; Brummer, T.; O’brien, P.M.; Daly, R.J. Dual-Specificity Phosphatases: Critical Regulators with Diverse Cellular Targets. Biochem. J. 2009, 418, 475–489. [Google Scholar] [CrossRef]
- Farooq, A.; Zhou, M.-M. Structure and Regulation of MAPK Phosphatases. Cell Signal 2004, 16, 769–779. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Tan, T.-H. DUSPs, to MAP Kinases and Beyond. Cell Biosci. 2012, 2, 24. [Google Scholar] [CrossRef]
- Ma, R.Y.M.; Tong, T.H.K.; Cheung, A.M.S.; Tsang, A.C.C.; Leung, W.Y.; Yao, K.-M. Raf/MEK/MAPK Signaling Stimulates the Nuclear Translocation and Transactivating Activity of FOXM1c. J. Cell Sci. 2005, 118, 795–806. [Google Scholar] [CrossRef]
- Brondello, J.-M.; Pouysségur, J.; McKenzie, F.R. Reduced MAP Kinase Phosphatase-1 Degradation after P42/P44MAPK-Dependent Phosphorylation. Science 1999, 286, 2514–2517. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Yang, J.-L. Cooperation of ERK and SCFSkp2 for MKP-1 Destruction Provides a Positive Feedback Regulation of Proliferating Signaling. J. Biol. Chem. 2006, 281, 915–926. [Google Scholar] [CrossRef]
- Cáceres, A.; Perdiguero, B.; Gómez, C.E.; Cepeda, M.V.; Caelles, C.; Sorzano, C.O.; Esteban, M. Involvement of the Cellular Phosphatase DUSP1 in Vaccinia Virus Infection. PLoS Pathog. 2013, 9, e1003719. [Google Scholar] [CrossRef]
- Choi, J.E.; Kwon, J.H.; Kim, J.-H.; Hur, W.; Sung, P.S.; Choi, S.W.; Yoon, S.K. Suppression of Dual Specificity Phosphatase I Expression Inhibits Hepatitis C Virus Replication. PLoS ONE 2015, 10, e0119172. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, A.C.; Caron, E.; Zucchini, N.; Mukawera, E.; Adam, D.; Mariani, M.K.; Gélinas, A.; Fortin, A.; Brochiero, E.; Grandvaux, N. DUSP1 Regulates Apoptosis and Cell Migration, but Not the JIP1-Protected Cytokine Response, during Respiratory Syncytial Virus and Sendai Virus Infection. Sci. Rep. 2017, 7, 17388. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. Coronaviruses Induce the Expression of the Dual Specificity Phosphatase DUSP1/MKP1. OSF Preprints 2020. [Google Scholar] [CrossRef]
- Chi, H.; Barry, S.P.; Roth, R.J.; Wu, J.J.; Jones, E.A.; Bennett, A.M.; Flavell, R.A. Dynamic Regulation of Pro- and Anti-Inflammatory Cytokines by MAPK Phosphatase 1 (MKP-1) in Innate Immune Responses. Proc. Natl. Acad. Sci. USA 2006, 103, 2274–2279. [Google Scholar] [CrossRef] [PubMed]
- Vacante, D.A.; Traub, R.; Major, E.O. Extension of JC Virus Host Range to Monkey Cells by Insertion of a Simian Virus 40 Enhancer into the JC Virus Regulatory Region. Virology 1989, 170, 353–361. [Google Scholar] [CrossRef]
- Nelson, C.D.S.; Carney, D.W.; Derdowski, A.; Lipovsky, A.; Gee, G.V.; O’Hara, B.; Williard, P.; DiMaio, D.; Sello, J.K.; Atwood, W.J. A Retrograde Trafficking Inhibitor of Ricin and Shiga-Like Toxins Inhibits Infection of Cells by Human and Monkey Polyomaviruses. mBio 2013, 4, e00729-13. [Google Scholar] [CrossRef]
- DuShane, J.K.; Wilczek, M.P.; Crocker, M.A.; Maginnis, M.S. High-Throughput Characterization of Viral and Cellular Protein Expression Patterns During JC Polyomavirus Infection. Front. Microbiol. 2019, 10, 783. [Google Scholar] [CrossRef]
- Mayberry, C.L.; Wilczek, M.P.; Fong, T.M.; Nichols, S.L.; Maginnis, M.S. GRK2 Mediates β-Arrestin Interactions with 5-HT 2 Receptors for JC Polyomavirus Endocytosis. J. Virol. 2021, 95, e02139-20. [Google Scholar] [CrossRef] [PubMed]
- Mayberry, C.L.; Soucy, A.N.; Lajoie, C.R.; DuShane, J.K.; Maginnis, M.S. JC Polyomavirus Entry by Clathrin-Mediated Endocytosis Is Driven by β-Arrestin. J. Virol. 2019, 93, e01948-18. [Google Scholar] [CrossRef]
- Shen, J.; Xing, W.; Liu, R.; Zhang, Y.; Xie, C.; Gong, F. MiR-32-5p Influences High Glucose-Induced Cardiac Fibroblast Proliferation and Phenotypic Alteration by Inhibiting DUSP1. BMC Mol. Biol. 2019, 20, 21. [Google Scholar] [CrossRef]
- Chapagain, M.L.; Verma, S.; Mercier, F.; Yanagihara, R.; Nerurkar, V.R. Polyomavirus JC Infects Human Brain Microvascular Endothelial Cells Independent of Serotonin Receptor 2A. Virology 2007, 364, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein–Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2020, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2020, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R Package for Visually Combining Expression Data with Functional Analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2, Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar] [CrossRef]
- Boveia, V.; Schutz-Geschwender, A. Detection of Blotted Proteins, Methods and Protocols. Methods Mol. Biol. 2015, 1314, 115–130. [Google Scholar] [CrossRef]
- Hoffman, G.R.; Moerke, N.J.; Hsia, M.; Shamu, C.E.; Blenis, J. A High-Throughput, Cell-Based Screening Method for SiRNA and Small Molecule Inhibitors of MTORC1 Signaling Using the In Cell Western Technique. Assay Drug Dev. Techn. 2010, 8, 186–199. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hillig, R.C.; Sautier, B.; Schroeder, J.; Moosmayer, D.; Hilpmann, A.; Stegmann, C.M.; Werbeck, N.D.; Briem, H.; Boemer, U.; Weiske, J.; et al. Discovery of Potent SOS1 Inhibitors That Block RAS Activation via Disruption of the RAS–SOS1 Interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 201812963. [Google Scholar] [CrossRef]
- Poltorak, M.; Meinert, I.; Stone, J.C.; Schraven, B.; Simeoni, L. Sos1 Regulates Sustained TCR-mediated Erk Activation. Eur. J. Immunol. 2014, 44, 1535–1540. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Hahn, W.C. Involvement of PP2A in Viral and Cellular Transformation. Oncogene 2005, 24, 7746–7755. [Google Scholar] [CrossRef]
- Bollag, B.; Hofstetter, C.A.; Reviriego-Mendoza, M.M.; Frisque, R.J. JC Virus Small t Antigen Binds Phosphatase PP2A and Rb Family Proteins and Is Required for Efficient Viral DNA Replication Activity. PLoS ONE 2010, 5, e10606. [Google Scholar] [CrossRef]
- Sariyer, I.K.; Khalili, K.; Safak, M. Dephosphorylation of JC Virus Agnoprotein by Protein Phosphatase 2A: Inhibition by Small t Antigen. Virology 2008, 375, 464–479. [Google Scholar] [CrossRef]
- Wlodarchak, N.; Xing, Y. PP2A as a Master Regulator of the Cell Cycle. Crit. Rev. Biochem. Mol. 2016, 51, 162–184. [Google Scholar] [CrossRef]
- Salojin, K.V.; Owusu, I.B.; Millerchip, K.A.; Potter, M.; Platt, K.A.; Oravecz, T. Essential Role of MAPK Phosphatase-1 in the Negative Control of Innate Immune Responses. J. Immunol. 2006, 176, 1899–1907. [Google Scholar] [CrossRef]
- Hammer, M.; Mages, J.; Dietrich, H.; Servatius, A.; Howells, N.; Cato, A.C.B.; Lang, R. Dual Specificity Phosphatase 1 (DUSP1) Regulates a Subset of LPS-Induced Genes and Protects Mice from Lethal Endotoxin Shock. J. Exp. Med. 2006, 203, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Sempere, M.; Chattopadhyay, S.; Rovira, A.; Rodriguez-Fanjul, V.; Belda-Iniesta, C.; Tapia, M.; Cejas, P.; Machado-Pinilla, R.; Manguan-García, C.; Sánchez-Pérez, I.; et al. MKP1 Repression Is Required for the Chemosensitizing Effects of NF-ΚB and PI3K Inhibitors to Cisplatin in Non-Small Cell Lung Cancer. Cancer Lett. 2009, 286, 206–216. [Google Scholar] [CrossRef]
- Barres, B.A. The Mystery and Magic of Glia: A Perspective on Their Roles in Health and Disease. Neuron 2008, 60, 430–440. [Google Scholar] [CrossRef]
- Choi, S.S.; Lee, H.J.; Lim, I.; Satoh, J.; Kim, S.U. Human Astrocytes: Secretome Profiles of Cytokines and Chemokines. PLoS ONE 2014, 9, e92325. [Google Scholar] [CrossRef]
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and Pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef] [PubMed]
- Schmierer, B.; Hill, C.S. TGFβ–SMAD Signal Transduction: Molecular Specificity and Functional Flexibility. Nat. Rev. Mol. Cell Bio. 2007, 8, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, V.; Jensen, P.N.; Major, E.O. MEK1/2 Inhibitors Block Basal and Transforming Growth Factor Β1-Stimulated JC Virus Multiplication. J. Virol. 2007, 81, 6412–6418. [Google Scholar] [CrossRef][Green Version]
- Rohini, M.; Arumugam, B.; Vairamani, M.; Selvamurugan, N. Stimulation of ATF3 Interaction with Smad4 via TGF-Β1 for Matrix Metalloproteinase 13 Gene Activation in Human Breast Cancer Cells. Int. J. Biol. Macromol. 2019, 134, 954–961. [Google Scholar] [CrossRef] [PubMed]





| Protein | 1⁰ Antibody (Dilution, Manufacturer) | 2⁰ Antibody (Dilution, Manufacturer) |
|---|---|---|
| JCPyV T Ag | PAB962 (1:5, hybridoma) | anti-mouse Alexa Fluor 594 (1:1000, Thermo Fisher Scientific) |
| Total ERK (p44/42 MAPK) | 4695S (1:500, CST) | anti-rabbit IRDye 800CW (1:10,000, LI-COR) |
| Phospho-p44/42 MAPK (ERK1/2) (T202/Y204) | 9101S (1:750, CST) | |
| Total DUSP1 | sc-373841 (1:75, Santa Cruz Biotechnology) | anti-mouse IRDye 800CW (1:10,000, LI-COR) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilczek, M.P.; Armstrong, F.J.; Geohegan, R.P.; Mayberry, C.L.; DuShane, J.K.; King, B.L.; Maginnis, M.S. The MAPK/ERK Pathway and the Role of DUSP1 in JCPyV Infection of Primary Astrocytes. Viruses 2021, 13, 1834. https://doi.org/10.3390/v13091834
Wilczek MP, Armstrong FJ, Geohegan RP, Mayberry CL, DuShane JK, King BL, Maginnis MS. The MAPK/ERK Pathway and the Role of DUSP1 in JCPyV Infection of Primary Astrocytes. Viruses. 2021; 13(9):1834. https://doi.org/10.3390/v13091834
Chicago/Turabian StyleWilczek, Michael P., Francesca J. Armstrong, Remi P. Geohegan, Colleen L. Mayberry, Jeanne K. DuShane, Benjamin L. King, and Melissa S. Maginnis. 2021. "The MAPK/ERK Pathway and the Role of DUSP1 in JCPyV Infection of Primary Astrocytes" Viruses 13, no. 9: 1834. https://doi.org/10.3390/v13091834
APA StyleWilczek, M. P., Armstrong, F. J., Geohegan, R. P., Mayberry, C. L., DuShane, J. K., King, B. L., & Maginnis, M. S. (2021). The MAPK/ERK Pathway and the Role of DUSP1 in JCPyV Infection of Primary Astrocytes. Viruses, 13(9), 1834. https://doi.org/10.3390/v13091834





