Detection of Low-Copy Human Virus DNA upon Prolonged Formalin Fixation
Abstract
:1. Introduction
2. Materials and Methods
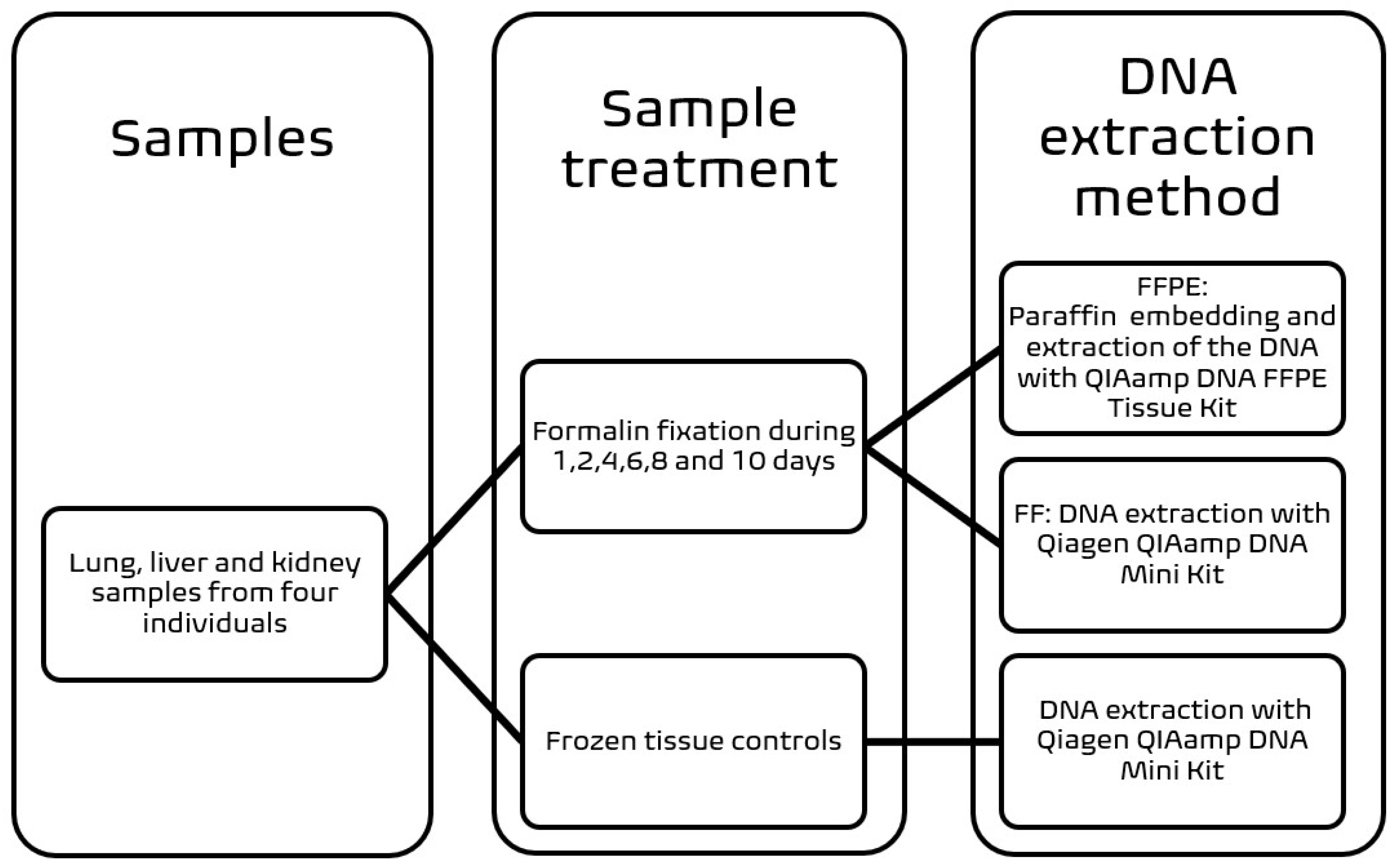
2.1. Sample Processing
2.2. DNA Extraction Methods and Quality of Total DNA
2.3. Quantitative PCRs
2.4. Library Preparation, Viral Enrichment, and DNA Sequencing
2.5. NGS Data Analysis
2.6. Statistical Analysis
3. Results
3.1. Total DNA Analysis
3.2. Viral DNA Detection in Frozen Controls and Formalin-Fixed Samples by qPCR
3.3. Viral Targeted Enrichment and NGS
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Do, H.; Dobrovic, A. Sequence Artifacts in DNA from Formalin-Fixed Tissues: Causes and Strategies for Minimization. Clin. Chem. 2015, 61, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Jackson, V. Studies on Histone Organization in the Nucleosome Using Formaldehyde as a Reversible Cross-Linking Agent. Cell 1978, 15, 945–954. [Google Scholar] [CrossRef]
- Vitošević, K.; Todorović, M.; Varljen, T.; Slović, Ž.; Matić, S.; Todorović, D. Effect of Formalin Fixation on Pcr Amplification of DNA Isolated from Healthy Autopsy Tissues. Acta Histochem. 2018, 120, 780–788. [Google Scholar] [CrossRef]
- Pyöriä, L.; Toppinen, M.; Mäntylä, E.; Hedman, L.; Aaltonen, L.M.; Vihinen-Ranta, M.; Ilmarinen, T.; Söderlund-Venermo, M.; Hedman, K.; Perdomo, M.F. Extinct Type of Human Parvovirus B19 Persists in Tonsillar B Cells. Nat. Commun. 2017, 8, 14930. [Google Scholar] [CrossRef]
- Toppinen, M.; Sajantila, A.; Pratas, D.; Hedman, K.; Perdomo, M.F. The Human Bone Marrow Is Host to the DNAs of Several Viruses. Front. Cell. Infect. Microbiol. 2021, 11, 329. [Google Scholar] [CrossRef]
- Toppinen, M.; Pratas, D.; Väisänen, E.; Söderlund-Venermo, M.; Hedman, K.; Perdomo, M.F.; Sajantila, A. The Landscape of Persistent Human DNA Viruses in Femoral Bone. Forensic Sci. Int. Genet. 2020, 48, 102353. [Google Scholar] [CrossRef]
- Lecuit, M.; Eloit, M. The Diagnosis of Infectious Diseases by Whole Genome next Generation Sequencing: A New Era Is Opening. Front. Cell. Infect. Microbiol. 2014, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- McDonough, S.J.; Bhagwate, A.; Sun, Z.; Wang, C.; Zschunke, M.; Gorman, J.A.; Kopp, K.J.; Cunningham, J.M. Use of FFPE-Derived DNA in next Generation Sequencing: DNA Extraction Methods. PLoS ONE 2019, 14, e0211400. [Google Scholar] [CrossRef] [Green Version]
- Bonnet, E.; Moutet, M.-L.; Baulard, C.; Bacq-Daian, D.; Sandron, F.; Mesrob, L.; Fin, B.; Delépine, M.; Palomares, M.-A.; Jubin, C.; et al. Performance Comparison of Three DNA Extraction Kits on Human Whole-Exome Data from Formalin-Fixed Paraffin-Embedded Normal and Tumor Samples. PLoS ONE 2018, 13, e0195471. [Google Scholar] [CrossRef] [Green Version]
- Ambulos, N.P.; Schumaker, L.M.; Mathias, T.J.; White, R.; Troyer, J.; Wells, D.; Cullen, K.J. Next-Generation Sequencing-Based HPV Genotyping Assay Validated in Formalin-Fixed, Paraffin-Embedded Oropharyngeal and Cervical Cancer Specimens. J. Biomol. Tech. 2016, 27, 46–52. [Google Scholar] [CrossRef] [Green Version]
- Arvia, R.; Sollai, M.; Pierucci, F.; Urso, C.; Massi, D.; Zakrzewska, K. Droplet Digital PCR (DdPCR) vs Quantitative Real-Time PCR (QPCR) Approach for Detection and Quantification of Merkel Cell Polyomavirus (MCPyV) DNA in Formalin Fixed Paraffin Embedded (FFPE) Cutaneous Biopsies. J. Virol. Methods 2017, 246, 15–20. [Google Scholar] [CrossRef]
- Božić, L.; Jovanović, T.; Šmitran, A.; Janković, M.; Knežević, A. Comparison of HPV Detection Rate in Formalin-fixed Paraffin-embedded Tissues of Head and Neck Carcinoma Using Two DNA Extraction Kits and Three Amplification Methods. Eur. J. Oral Sci. 2020, 128, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Lagheden, C.; Eklund, C.; Kleppe, S.N.; Unger, E.R.; Dillner, J.; Sundström, K. Validation of a Standardized Extraction Method for Formalin-Fixed Paraffin-Embedded Tissue Samples. J. Clin. Virol. 2016, 80, 36–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollo, F.; Donà, M.G.; Pichi, B.; Pellini, R.; Covello, R.; Benevolo, M. Evaluation of the Anyplex II HPV28 Assay in the Detection of Human Papillomavirus in Archival Samples of Oropharyngeal Carcinomas. Arch. Pathol. Lab. Med. 2020, 144, 620–625. [Google Scholar] [CrossRef] [Green Version]
- Swan, D.C.; Tucker, R.A.; Tortolero-Luna, G.; Mitchell, M.F.; Wideroff, L.; Unger, E.R.; Nisenbaum, R.A.; Reeves, W.C.; Icenogle, J.P. Human Papillomavirus (HPV) DNA Copy Number Is Dependent on Grade of Cervical Disease and HPV Type. J. Clin. Microbiol. 1999, 37, 1030–1034. [Google Scholar] [CrossRef] [Green Version]
- Toppinen, M.; Norja, P.; Aaltonen, L.M.; Wessberg, S.; Hedman, L.; Söderlund-Venermo, M.; Hedman, K. A New Quantitative PCR for Human Parvovirus B19 Genotypes. J. Virol. Methods 2015, 218, 40–45. [Google Scholar] [CrossRef]
- Pyöriä, L.; Jokinen, M.; Toppinen, M.; Salminen, H.; Vuorinen, T.; Hukkanen, V.; Schmotz, C.; Elbasani, E.; Ojala, P.M.; Hedman, K.; et al. HERQ-9 Is a New Multiplex PCR for Differentiation and Quantification of All Nine Human Herpesviruses. mSphere 2020, 5, e00265-20. [Google Scholar] [CrossRef]
- Pratas, D.; Toppinen, M.; Pyoria, L.; Hedman, K.; Sajantila, A.; Perdomo, M.F. A Hybrid Pipeline for Reconstruction and Analysis of Viral Genomes at Multi-Organ Level. GigaScience 2020, 9, giaa086. [Google Scholar] [CrossRef]
- Almeida, J.R.; Pinho, A.J.; Oliveira, J.L.; Fajarda, O.; Pratas, D. GTO: A Toolkit to Unify Pipelines in Genomic and Proteomic Research. SoftwareX 2020, 12, 100535. [Google Scholar] [CrossRef]
- Pratas, D.; Hosseini, M.; Grilo, G.; Pinho, A.J.; Silva, R.M.; Caetano, T.; Carneiro, J.; Pereira, F. Metagenomic Composition Analysis of an Ancient Sequenced Polar Bear Jawbone from Svalbard. Genes 2018, 9, 445. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Klenner, J.; Kohl, C.; Dabrowski, P.W.; Nitsche, A. Comparing Viral Metagenomic Extraction Methods. Curr. Issues Mol. Biol. 2017, 24, 59–70. [Google Scholar] [CrossRef]
- Gilbert, M.T.P.; Haselkorn, T.; Bunce, M.; Sanchez, J.J.; Lucas, S.B.; Jewell, L.D.; van Marck, E.; Worobey, M. The Isolation of Nucleic Acids from Fixed, Paraffin-Embedded Tissues-Which Methods Are Useful When? PLoS ONE 2007, 2, e537. [Google Scholar] [CrossRef]
- Robbe, P.; Popitsch, N.; Knight, S.J.L.; Antoniou, P.; Becq, J.; He, M.; Kanapin, A.; Samsonova, A.; Vavoulis, D.V.; Ross, M.T.; et al. Clinical Whole-Genome Sequencing from Routine Formalin-Fixed, Paraffin-Embedded Specimens: Pilot Study for the 100,000 Genomes Project. Genet. Med. 2018, 20, 1196–1205. [Google Scholar] [CrossRef] [Green Version]
- Kocjan, B.J.; Hošnjak, L.; Poljak, M. Detection of Alpha Human Papillomaviruses in Archival Formalin-Fixed, Paraffin-Embedded (FFPE) Tissue Specimens. J. Clin. Virol. 2016, 76, S88–S97. [Google Scholar] [CrossRef]
- Reid, K.M.; Maistry, S.; Ramesar, R.; Heathfield, L.J. A Review of the Optimisation of the Use of Formalin Fixed Paraffin Embedded Tissue for Molecular Analysis in a Forensic Post-Mortem Setting. Forensic Sci. Int. 2017, 280, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Cummings, M.; King, H.; Hurst, J.; Tanner, G.; Khazin, L.; Thompson, P.; Gray, A.; Gahir, N.; Cartlidge, C.; Farooq, Z.; et al. Decreasing Formalin Concentration Improves Quality of DNA Extracted from Formalin-Fixed Paraffin-Embedded Tissue Specimens without Compromising Tissue Morphology or Immunohistochemical Staining. J. Clin. Pathol. 2020, 73, 514–518. [Google Scholar] [CrossRef]
- Legrand, B.; de Mazancourt, P.; Durigon, M.; Khalifat, V.; Crainic, K. DNA Genotyping of Unbuffered Formalin Fixed Paraffin Embedded Tissues. Forensic Sci. Int. 2002, 125, 205–211. [Google Scholar] [CrossRef]
- Carpenter, M.L.; Buenrostro, J.D.; Valdiosera, C.; Schroeder, H.; Allentoft, M.E.; Sikora, M.; Rasmussen, M.; Gravel, S.; Guillén, S.; Nekhrizov, G.; et al. Pulling out the 1%: Whole-Genome Capture for the Targeted Enrichment of Ancient Dna Sequencing Libraries. Am. J. Hum. Genet. 2013, 93, 852–864. [Google Scholar] [CrossRef] [Green Version]
- Duncavage, E.J.; Magrini, V.; Becker, N.; Armstrong, J.R.; Demeter, R.T.; Wylie, T.; Abel, H.J.; Pfeifer, J.D. Hybrid Capture and Next-Generation Sequencing Identify Viral Integration Sites from Formalin-Fixed, Paraffin-Embedded Tissue. J. Mol. Diagn. 2011, 13, 325–333. [Google Scholar] [CrossRef]
- Bodewes, R.; van Run, P.R.W.A.; Schürch, A.C.; Koopmans, M.P.G.; Osterhaus, A.D.M.E.; Baumgärtner, W.; Kuiken, T.; Smits, S.L. Virus Characterization and Discovery in Formalin-Fixed Paraffin-Embedded Tissues. J. Virol. Methods 2015, 214, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, F.; Bennett, S.; Modha, S.; Murdoch, D.; Gunson, R.; Murcia, P.R. The Use of next Generation Sequencing in the Diagnosis and Typing of Respiratory Infections. J. Clin. Virol. 2015, 69, 96–100. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, D.; Uhl, B.; Sailer, V.; Holmes, E.E.; Jung, M.; Meller, S.; Kristiansen, G. Improved PCR Performance Using Template DNA from Formalin-Fixed and Paraffin-Embedded Tissues by Overcoming PCR Inhibition. PLoS ONE 2013, 8, e77771. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Reid, A.H.; Krafft, A.E.; Bijwaard, K.E.; Fanning, T.G. Initial Genetic Characterization of the 1918 “Spanish” Influenza Virus. Science 1997, 275, 1793–1796. [Google Scholar] [CrossRef] [Green Version]
- Düx, A.; Lequime, S.; Patrono, L.V.; Vrancken, B.; Boral, S.; Gogarten, J.F.; Hilbig, A.; Horst, D.; Merkel, K.; Prepoint, B.; et al. Measles Virus and Rinderpest Virus Divergence Dated to the Sixth Century BCE. Science 2020, 368, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, D.W.; Zagordi, O.; Geissberger, F.D.; Kufner, V.; Schmutz, S.; Böni, J.; Metzner, K.J.; Trkola, A.; Huber, M. Optimization and Validation of Sample Preparation for Metagenomic Sequencing of Viruses in Clinical Samples. Microbiome 2017, 5, 94. [Google Scholar] [CrossRef] [Green Version]





| Individual | Tissue | EBV | B19V | TTV | HHV-6B | HHV-7 |
|---|---|---|---|---|---|---|
| 1 | Lung | 114 | 27.5 | |||
| Liver | ||||||
| Kidney | 165 | 21.6 | ||||
| 2 | Lung | 472 | 43.6 | |||
| Liver | 911 | |||||
| Kidney | 101 | 25.4 | 5.79 | |||
| 3 | Lung | 691 | 45.7 | |||
| Liver | 50.3 | 249 | ||||
| Kidney | 227 | 57.7 | ||||
| 4 | Lung | 14.6 | 536 | 154 | 242 | 23.5 |
| Liver | 1100 | 1600 | 4720 | 39.4 | ||
| Kidney | 14.5 | 2970 | 5.97 | 3370 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mielonen, O.I.; Pratas, D.; Hedman, K.; Sajantila, A.; Perdomo, M.F. Detection of Low-Copy Human Virus DNA upon Prolonged Formalin Fixation. Viruses 2022, 14, 133. https://doi.org/10.3390/v14010133
Mielonen OI, Pratas D, Hedman K, Sajantila A, Perdomo MF. Detection of Low-Copy Human Virus DNA upon Prolonged Formalin Fixation. Viruses. 2022; 14(1):133. https://doi.org/10.3390/v14010133
Chicago/Turabian StyleMielonen, Outi I., Diogo Pratas, Klaus Hedman, Antti Sajantila, and Maria F. Perdomo. 2022. "Detection of Low-Copy Human Virus DNA upon Prolonged Formalin Fixation" Viruses 14, no. 1: 133. https://doi.org/10.3390/v14010133
APA StyleMielonen, O. I., Pratas, D., Hedman, K., Sajantila, A., & Perdomo, M. F. (2022). Detection of Low-Copy Human Virus DNA upon Prolonged Formalin Fixation. Viruses, 14(1), 133. https://doi.org/10.3390/v14010133







