Chimeric Virus-like Particles Co-Displaying Hemagglutinin Stem and the C-Terminal Fragment of DnaK Confer Heterologous Influenza Protection in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Lines and Influenza Viruses
2.3. Construction and Expression of the Chimeric VLPs
2.4. Transmission Electron Microscopy
2.5. In Vitro Assays for Inflammation Responses
2.6. Animal Immunization and Challenge Studies
2.7. Determination of Specific IgG Antibody Titers
2.8. Determination of Lung Viral Load
2.9. ELISPOT Assay
2.10. Serum Microneutralization Assay
2.11. Statistical Analysis
3. Results
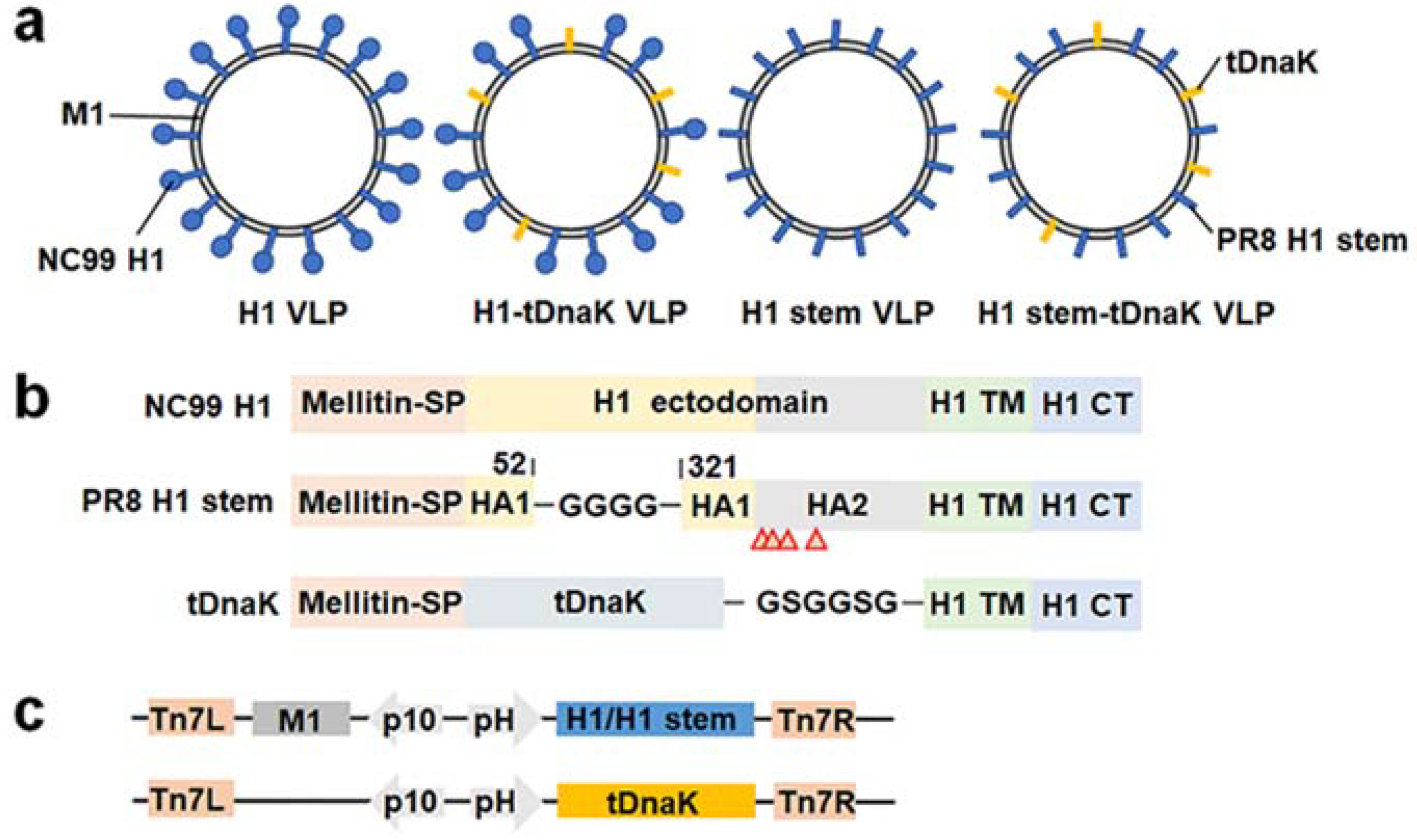
3.1. Construction and Characterization of Influenza Chimeric VLPs

3.2. The cVLPs Induced Humoral and Cellular Immune Responses
3.3. Immunizations of the cVLPs Conferred Heterologous Influenza Protection against Lethal-Dose Infections
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Becker, T.; Elbahesh, H.; Reperant, L.A.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E. Influenza Vaccines: Successes and Continuing Challenges. J. Infect. Dis. 2021, 224, S405–S419. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Wei, C.J.; Crank, M.C.; Shiver, J.; Graham, B.S.; Mascola, J.R.; Nabel, G.J. Next-generation influenza vaccines: Opportunities and challenges. Nat. Rev. Drug Discov. 2020, 19, 239–252. [Google Scholar] [CrossRef]
- Kosik, I.; Yewdell, J.W. Influenza Hemagglutinin and Neuraminidase: Yin–Yang Proteins Coevolving to Thwart Immunity. Viruses 2019, 11, 346. [Google Scholar] [CrossRef]
- Kosik, I.; Ince, W.L.; Gentles, L.E.; Oler, A.J.; Kosikova, M.; Angel, M.; Magadan, J.G.; Xie, H.; Brooke, C.B.; Yewdell, J.W. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018, 14, e1006796. [Google Scholar]
- Fulton, B.O.; Sun, W.; Heaton, N.S.; Palese, P. The Influenza B Virus Hemagglutinin Head Domain Is Less Tolerant to Transposon Mutagenesis than That of the Influenza A Virus. J. Virol. 2018, 92, e00754-18. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly Conserved Protective Epitopes on Influenza B Viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef]
- Wu, Y.; Cho, M.; Shore, D.; Song, M.; Choi, J.; Jiang, T.; Deng, Y.-Q.; Bourgeois, M.; Almli, L.; Yang, H.; et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 2015, 6, 7708. [Google Scholar] [CrossRef]
- Harshbarger, W.D.; Deming, D.; Lockbaum, G.J.; Attatippaholkun, N.; Kamkaew, M.; Hou, S.; Somasundaran, M.; Wang, J.P.; Finberg, R.W.; Zhu, Q.K.; et al. Unique structural solution from a VH3-30 antibody targeting the hemagglutinin stem of influenza A viruses. Nat. Commun. 2021, 12, 559. [Google Scholar] [CrossRef]
- Benton, D.J.; Nans, A.; Calder, L.J.; Turner, J.; Neu, U.; Lin, Y.P.; Ketelaars, E.; Kallewaard, N.L.; Corti, D.; Lanzavecchia, A.; et al. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 2018, 115, 10112–10117. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; Van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.-J.; Kanekiyo, M.; Kong, W.-P.; Gallagher, J.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Krammer, F.; Pica, N.; Hai, R.; Tan, G.S.; Palese, P. Hemagglutinin Stalk-Reactive Antibodies Are Boosted following Sequential Infection with Seasonal and Pandemic H1N1 Influenza Virus in Mice. J. Virol. 2012, 86, 10302–10307. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Guptill, J.; Naficy, A.; Nachbagauer, R.; Berlanda-Scorza, F.; Feser, J.; Wilson, P.C.; Solórzano, A.; Van Der Wielen, M.; Walter, E.B.; et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 2020, 20, 80–91. [Google Scholar] [CrossRef]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef]
- Nagai, K.; Domon, H.; Maekawa, T.; Oda, M.; Hiyoshi, T.; Tamura, H.; Yonezawa, D.; Arai, Y.; Yokoji, M.; Tabeta, K.; et al. Pneumococcal DNA-binding proteins released through autolysis induce the production of proinflammatory cytokines via toll-like receptor 4. Cell. Immunol. 2018, 325, 14–22. [Google Scholar] [CrossRef]
- Wang, Y.; Kelly, C.G.; Singh, M.; McGowan, E.G.; Carrara, A.-S.; Bergmeier, L.A.; Lehner, T. Stimulation of Th1-Polarizing Cytokines, C-C Chemokines, Maturation of Dendritic Cells, and Adjuvant Function by the Peptide Binding Fragment of Heat Shock Protein 70. J. Immunol. 2002, 169, 2422–2429. [Google Scholar] [CrossRef]
- Tobian, A.A.R.; Canaday, D.H.; Boom, W.H.; Harding, C.V. Bacterial Heat Shock Proteins Promote CD91-Dependent Class I MHC Cross-Presentation of Chaperoned Peptide to CD8+ T Cells by Cytosolic Mechanisms in Dendritic Cells versus Vacuolar Mechanisms in Macrophages. J. Immunol. 2004, 172, 5277–5286. [Google Scholar] [CrossRef]
- Ericsson, M.; Kroca, M.; Johansson, T.; Sjostedt, A.; Tarnvik, A. Long-Lasting Recall Response of CD4 and CD8 (infinity) b T Cells, but not gd T Cells, to Heat Shock Proteins of Francisella tularensis. Scand. J. Infect. Dis. 2001, 33, 145–152. [Google Scholar] [PubMed]
- Deng, L.; Ibañez, L.I.; Bossche, V.V.D.; Roose, K.; Youssef, S.A.; De Bruin, A.; Fiers, W.; Saelens, X. Protection against Influenza A Virus Challenge with M2e-Displaying Filamentous Escherichia coli Phages. PLoS ONE 2015, 10, e0126650. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, M.A.; Klotchenko, S.A.; Vasin, A.V. Development of a multiplex quantitative PCR assay for the analysis of human cytokine gene expression in influenza A virus-infected cells. J. Immunol. Methods 2016, 430, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Cha, M.; Jin, Y.-Z.; Park, J.W.; Lee, K.M.; Han, S.H.; Choi, B.S.; Lee, J.H. Three-dimensional printed polylactic acid scaffold integrated with BMP-2 laden hydrogel for precise bone regeneration. Biomater. Res. 2021, 25, 35. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Deng, L.; Mohan, T.; Chang, T.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.-M.; Kang, S.-M.; Compans, R.W.; Champion, J.A.; Wang, B.-Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef]
- Chávez-Sánchez, L.; Chávez-Rueda, K.; Legorreta-Haquet, M.V.; Zenteno, E.; Ledesma-Soto, Y.; Montoya-Díaz, E.; Tesoro-Cruz, E.; Madrid-Miller, A.; Blanco-Favela, F. The activation of CD14, TLR4, and TLR2 by mmLDL induces IL-1β, IL-6, and IL-10 secretion in human monocytes and macrophages. Lipids Health Dis. 2010, 9, 117. [Google Scholar] [CrossRef]
- Van De Veerdonk, F.L.; Joosten, L.A.B.; Shaw, P.J.; Smeekens, S.P.; Malireddi, R.; Van Der Meer, J.W.M.; Kullberg, B.-J.; Netea, M.G.; Kanneganti, T.-D. The inflammasome drives protective Th1 and Th17 cellular responses in disseminated candidiasis. Eur. J. Immunol. 2011, 41, 2260–2268. [Google Scholar] [CrossRef]
- Subbiah, J.; Oh, J.; Kim, K.-H.; Shin, C.-H.; Park, B.R.; Bhatnagar, N.; Seong, B.-L.; Wang, B.-Z.; Kang, S.-M. A chimeric thermostable M2e and H3 stalk-based universal influenza A virus vaccine. npj Vaccines 2022, 7, 1–15. [Google Scholar] [CrossRef]
- Song, Y.; Zhu, W.; Wang, Y.; Deng, L.; Ma, Y.; Dong, C.; Gonzalez, G.X.; Kim, J.; Wei, L.; Kang, S.-M. Layered protein nanoparticles containing influenza B HA stalk inducted sustained cross-protection against viruses spanning both viral lineages. Biomaterials 2022, 287, 121664. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Seder, R.A.; Ahmed, R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 2003, 4, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, D.G.; Omokanye, A.; Schön, K.; Wenzel, U.A.; Bernasconi, V.; Bemark, M.; Kolpe, A.; El Bakkouri, K.; Ysenbaert, T.; Deng, L.; et al. M2e-tetramer-specific memory CD4 T cells are broadly protective against influenza infection. Mucosal Immunol. 2017, 11, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Kim, J.R.; Chang, T.Z.; Zhang, H.; Mohan, T.; Champion, J.A.; Wang, B.-Z. Protein nanoparticle vaccine based on flagellin carrier fused to influenza conserved epitopes confers full protection against influenza A virus challenge. Virology 2017, 509, 82–89. [Google Scholar] [CrossRef]
- Ashtekar, A.R.; Zhang, P.; Katz, J.; Deivanayagam, C.C.S.; Rallabhandi, P.; Vogel, S.N.; Michalek, S.M. TLR4-mediated activation of dendritic cells by the heat shock protein DnaK from Francisella tularensis. J. Leukoc. Biol. 2008, 84, 1434–1446. [Google Scholar] [CrossRef]
- Dabaghian, M.; Latifi, A.M.; Tebianian, M.; Dabaghian, F.; Ebrahimi, S.M. A truncated C-terminal fragment of Mycobacterium tuberculosis HSP70 enhances cell-mediated immune response and longevity of the total IgG to influenza A virus M2e protein in mice. Antivir. Res. 2015, 120, 23–31. [Google Scholar] [CrossRef]





| Primers | Sequences (5′-3′) |
|---|---|
| IL-1β-F | AGCTGATGGCCCTAAACAGA |
| IL-1β-R | TGGTGGTCGGAGATTCGTAG |
| IL-6-F | CCACTCACCTCTTCAGAACG |
| IL-6-R | CATCTTTGGAAGGTTCAGGTTG |
| GAPDH-F | CTCCTCCTGTTCGACAGTCA |
| GAPDH-R | CGACCAAATCCGTTGACTCC |
| Serum Samples | Virus Subtypes | |
|---|---|---|
| NJ76 H1N1 | PR8 H1N1 | |
| NC99 H1 VLP | <2 | <2 |
| NC99 H1-tDnaK VLP | 4 | 8 |
| PR8 H1 stem VLP | <2 | <2 |
| PR8 H1 stem-tDnaK VLP | <2 | <2 |
| PBS | <2 | <2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-C.; Liu, D.-J.; Yue, X.-Y.; Zhong, X.-Q.; Wu, X.; Chang, H.-Y.; Wang, B.-Z.; Wan, M.-Y.; Deng, L. Chimeric Virus-like Particles Co-Displaying Hemagglutinin Stem and the C-Terminal Fragment of DnaK Confer Heterologous Influenza Protection in Mice. Viruses 2022, 14, 2109. https://doi.org/10.3390/v14102109
Liu C-C, Liu D-J, Yue X-Y, Zhong X-Q, Wu X, Chang H-Y, Wang B-Z, Wan M-Y, Deng L. Chimeric Virus-like Particles Co-Displaying Hemagglutinin Stem and the C-Terminal Fragment of DnaK Confer Heterologous Influenza Protection in Mice. Viruses. 2022; 14(10):2109. https://doi.org/10.3390/v14102109
Chicago/Turabian StyleLiu, Cui-Cui, De-Jian Liu, Xin-Yu Yue, Xiu-Qin Zhong, Xuan Wu, Hai-Yan Chang, Bao-Zhong Wang, Mu-Yang Wan, and Lei Deng. 2022. "Chimeric Virus-like Particles Co-Displaying Hemagglutinin Stem and the C-Terminal Fragment of DnaK Confer Heterologous Influenza Protection in Mice" Viruses 14, no. 10: 2109. https://doi.org/10.3390/v14102109






