Pin-Pointing the Key Hubs in the IFN-γ Pathway Responding to SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptome Datasets Selection
2.2. Bioinformatics Analyses
2.2.1. RNA-Seq Analysis
2.2.2. Microarray Analysis
2.3. Gene Correlation
2.4. Profiling of Immune Cell Type Abundance from Bulk Gene Expression Data Using CIBERSORT
2.5. Gene Set Variation Analysis (GSVA)
2.6. Cell Culture Conditions
2.7. Poly (I:C) Treatment
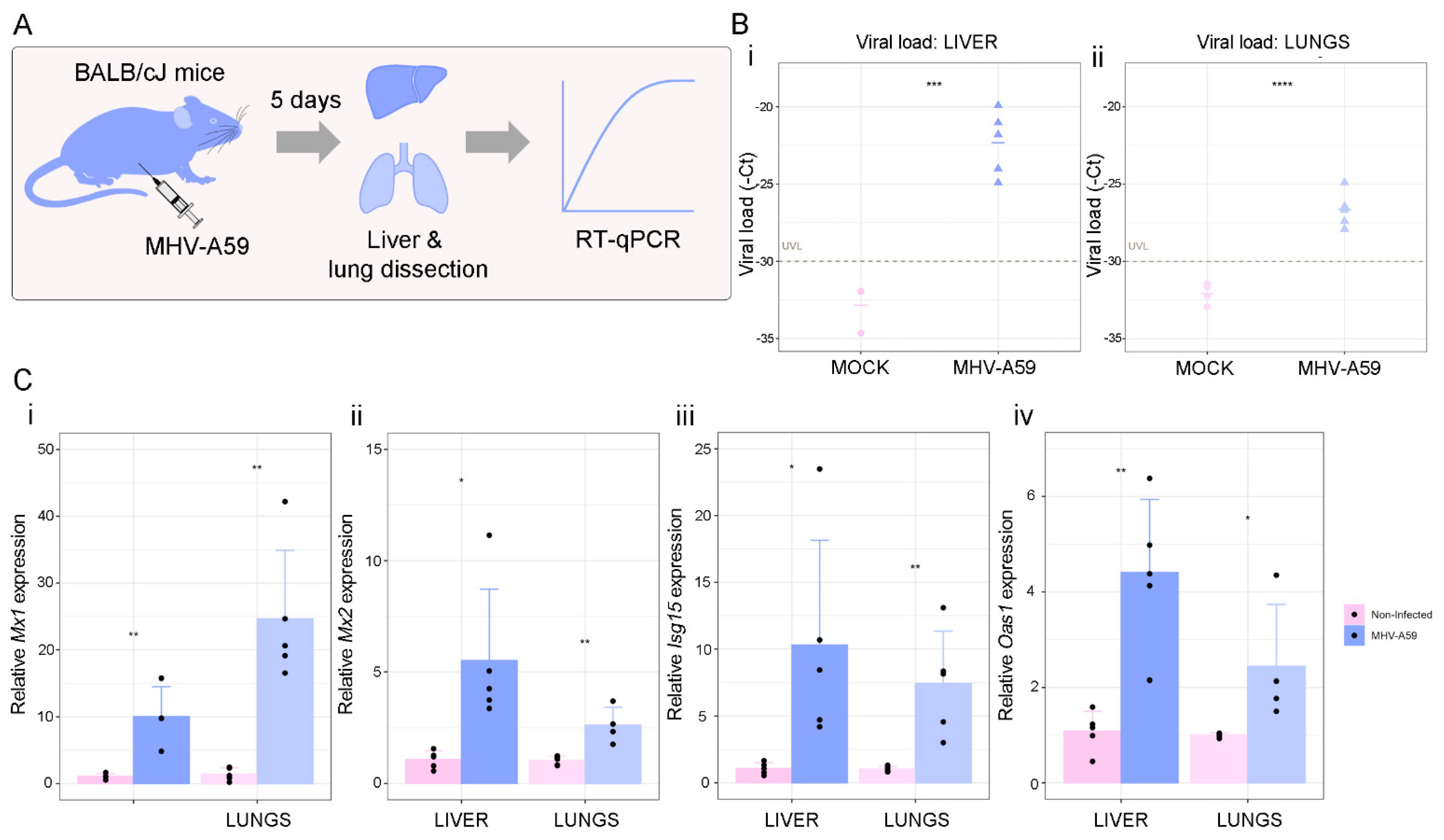
2.8. In Vivo Experiments
2.9. RNA Isolation, c-DNA Synthesis, and Quantitative Real-Time PCR (RT-qPCR)
2.10. Statistical Analyses
3. Results
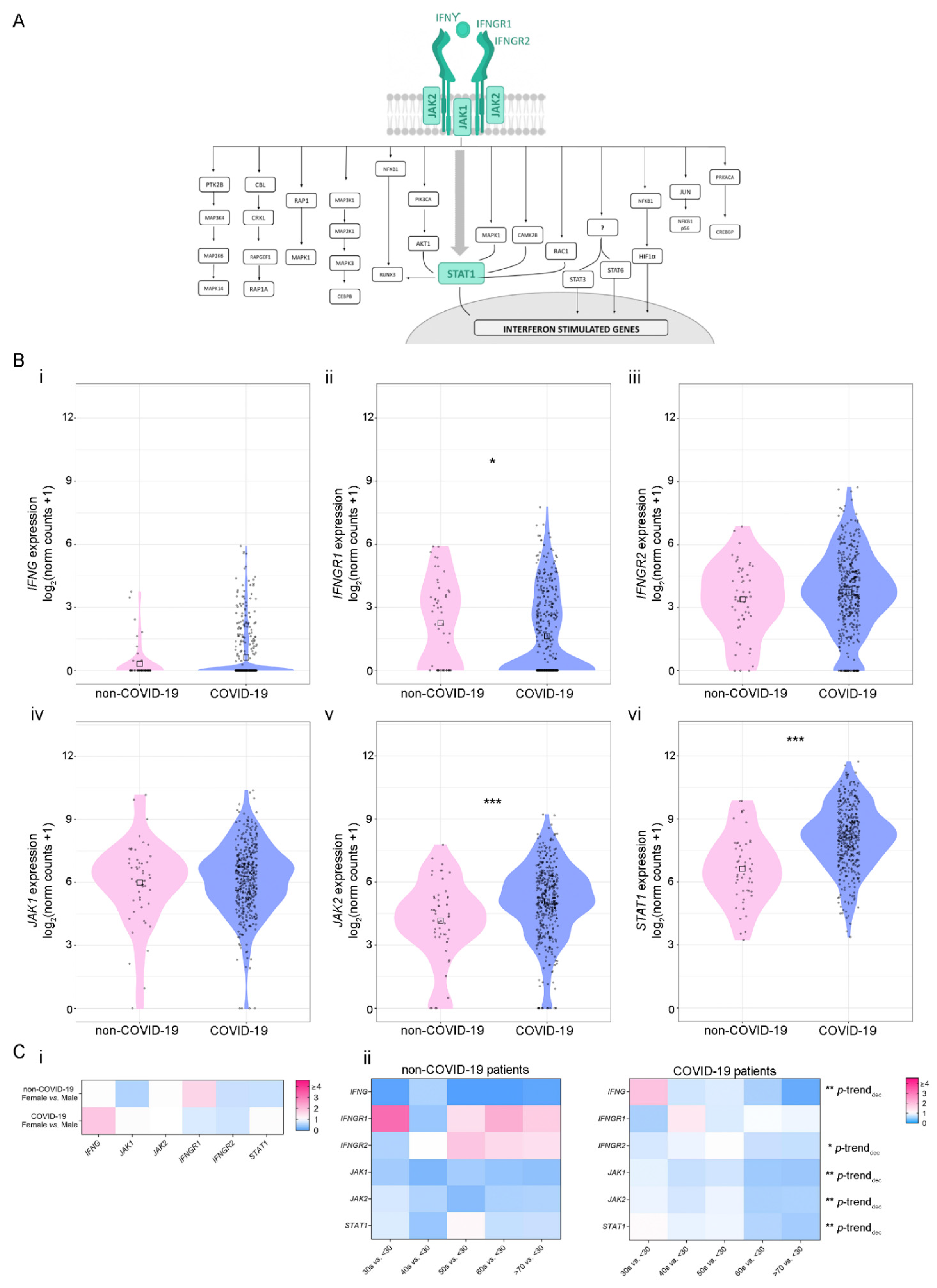
3.1. Analysis of IFN-γ Pathway in COVID-19-Positive and -Negative Patients
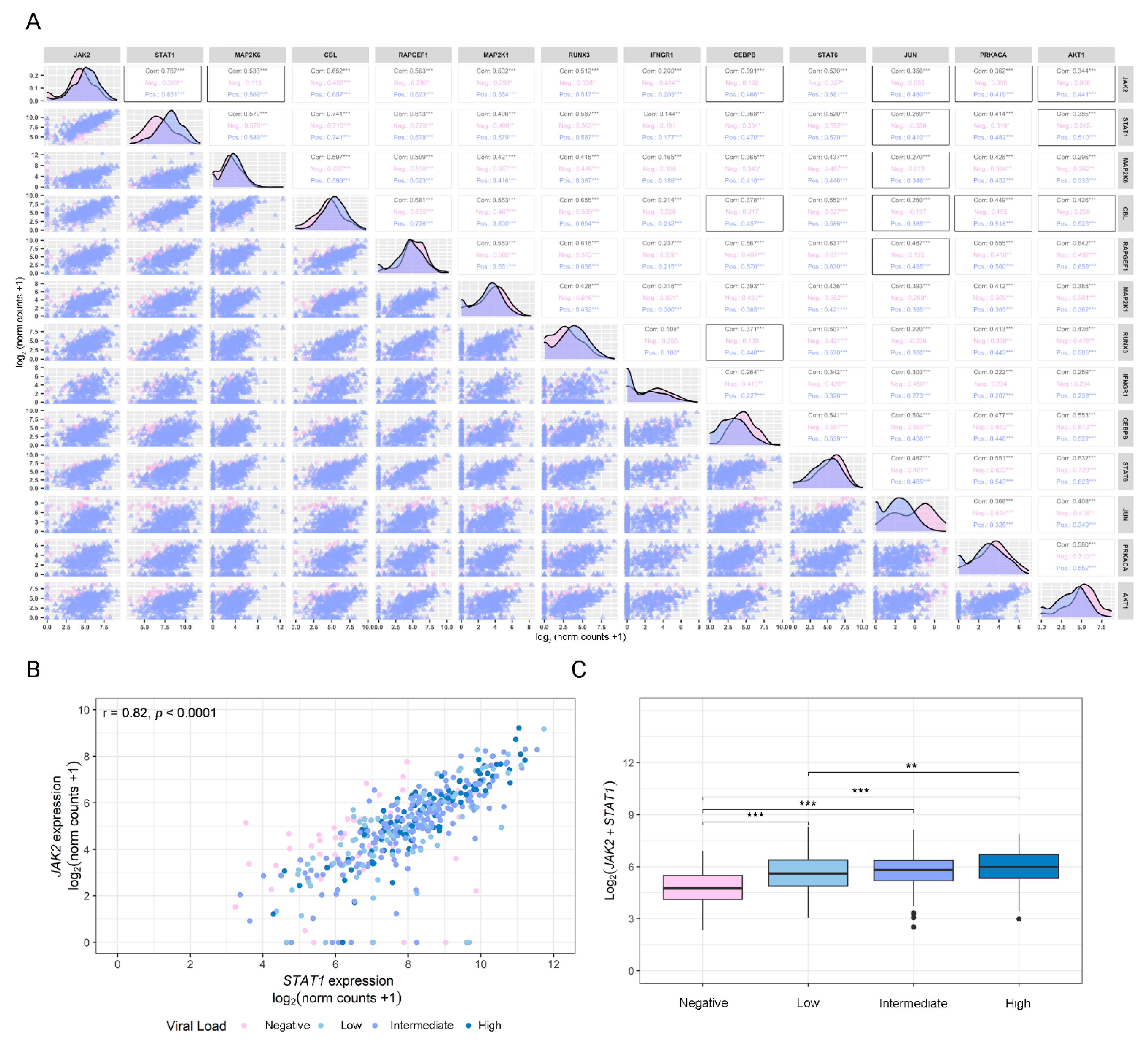
3.2. Gene Correlation Analysis of Dysregulated IFN-γ-Associated Genes in COVID-19-Positive and -Negative Patients
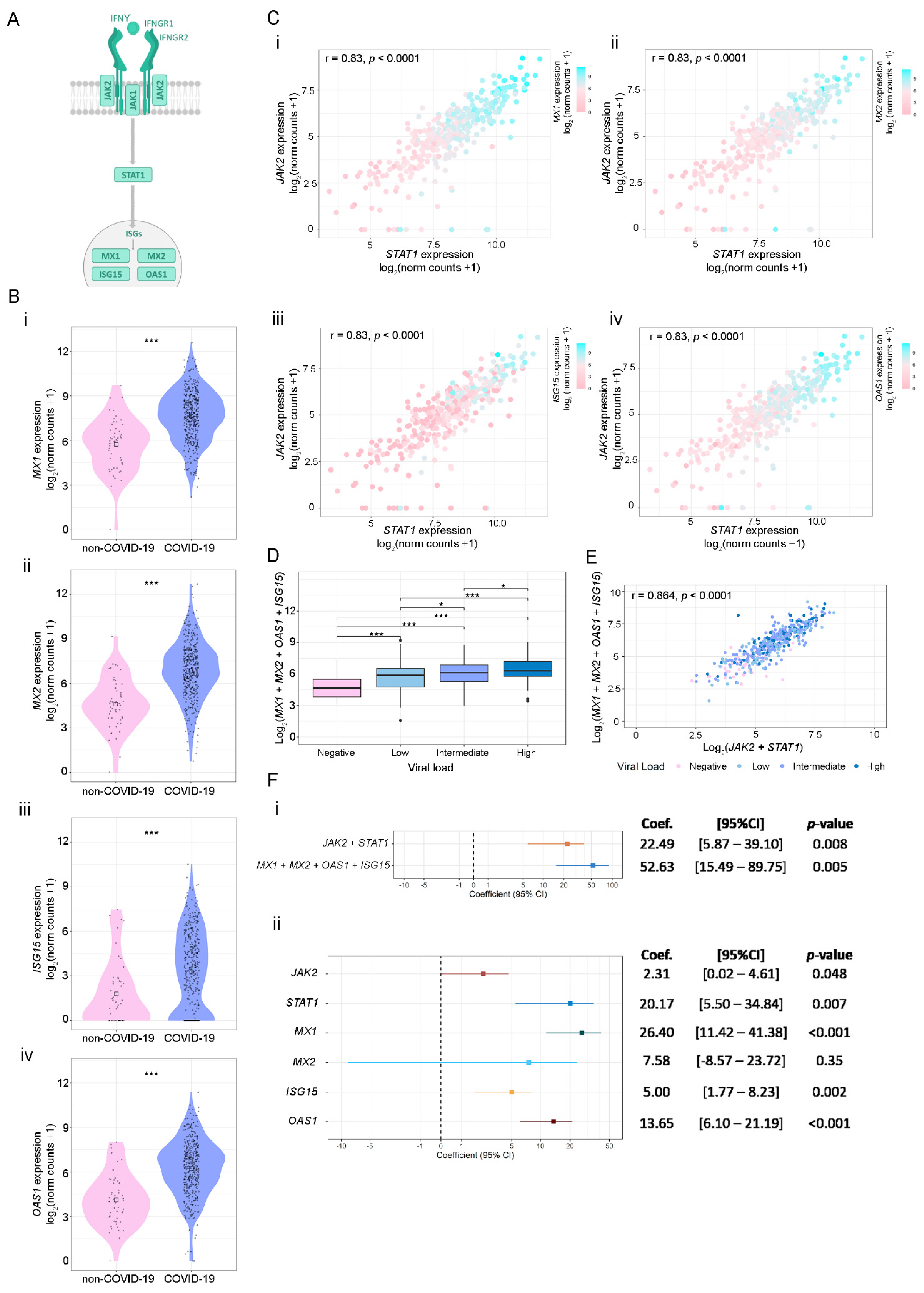
3.3. SARS-CoV-2 Viral Load Association with IFN-γ-Related Genes and ISGs
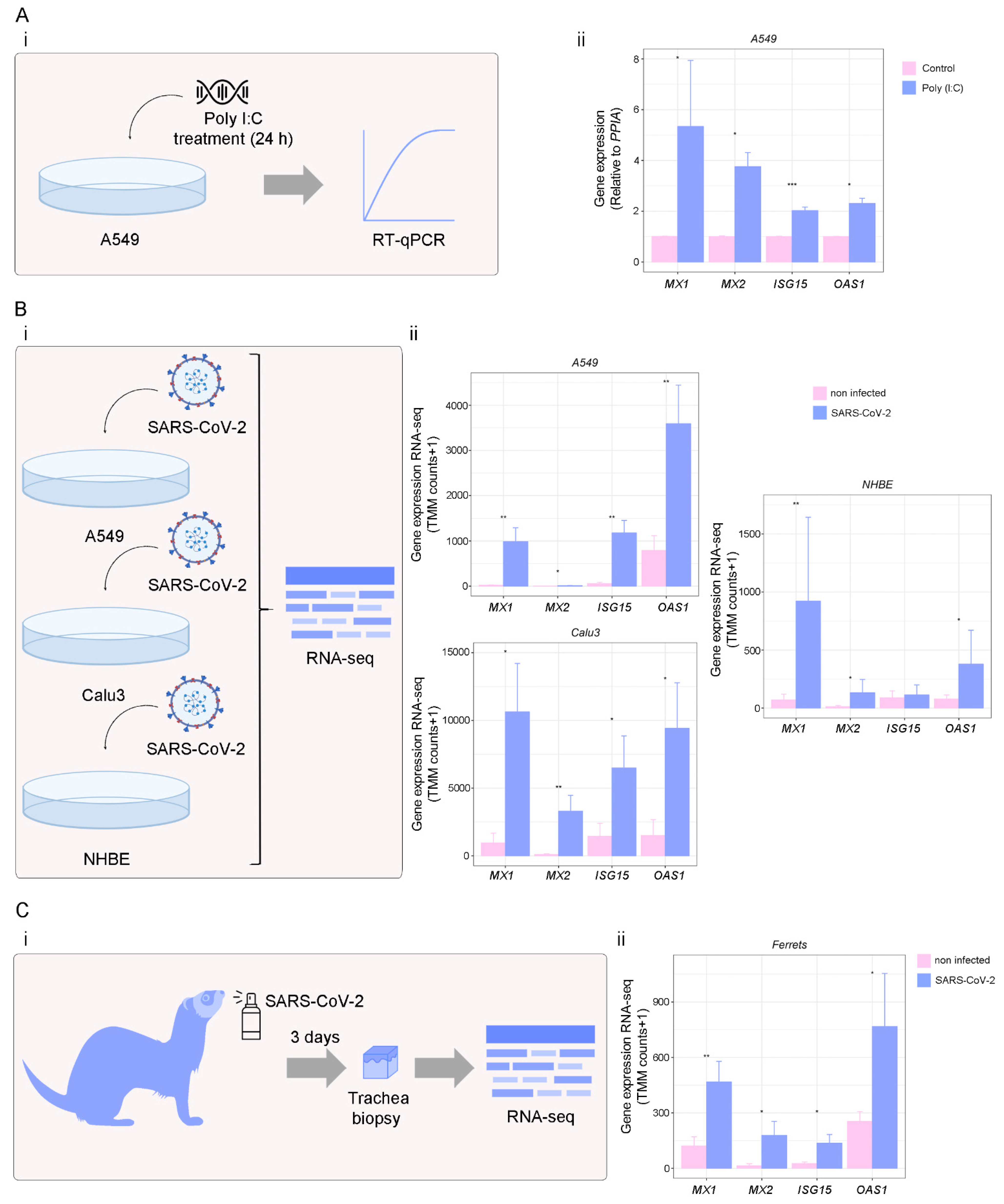
3.4. Targets of IFN-γ Pathway in Response to Viral Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meo, S.A.; Alhowikan, A.M.; Al-Khlaiwi, T.; Meo, I.M.; Halepoto, D.M.; Iqbal, M.; Usmani, A.M.; Hajjar, W.; Ahmed, N. Novel coronavirus 2019-nCoV: Prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2012–2019. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Guo, C.; Doldan, P.; Boulant, S. Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces. Front. Immunol. 2020, 11, 608645. [Google Scholar] [CrossRef] [PubMed]
- Palermo, E.; Di Carlo, D.; Sgarbanti, M.; Hiscott, J. Type I Interferons in COVID-19 Pathogenesis. Biology 2021, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Y.; Khan, N.; Close, B.J.; Goel, R.K.; Blum, B.; Tavares, A.H.; Kenney, D.; Conway, H.L.; Ewoldt, J.K.; Chitalia, V.C.; et al. SARS-CoV-2 Disrupts Proximal Elements in the JAK-STAT Pathway. J. Virol. 2021, 95, e0086221. [Google Scholar] [CrossRef] [PubMed]
- Meffre, E.; Iwasaki, A. Interferon deficiency can lead to severe COVID. Nature 2020, 587, 374–376. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; Hayday, A.C.; Rogers, A.J.; Towers, G.J.; Wack, A.; Zanoni, I. Anti-type I interferon antibodies as a cause of severe COVID-19. Fac. Rev. 2022, 11, 15. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Fukuda, Y.; Homma, T.; Inoue, H.; Onitsuka, C.; Ikeda, H.; Goto, Y.; Sato, Y.; Kimura, T.; Hirai, K.; Ohta, S.; et al. Downregulation of type III interferons in patients with severe COVID-19. J. Med. Virol. 2021, 93, 4559–4563. [Google Scholar] [CrossRef]
- Long, Q.-X.; Tang, X.-J.; Shi, Q.-L.; Li, Q.; Deng, H.-J.; Yuan, J.; Hu, J.-L.; Xu, W.; Zhang, Y.; Lv, F.-J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Lee, A.J.; Ashkar, A.A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef] [PubMed]
- Majoros, A.; Platanitis, E.; Kernbauer-Hölzl, E.; Rosebrock, F.; Müller, M.; Decker, T. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Koepke, L.; Kirchhoff, F.; Sparrer, K.M.J. Interferon antagonists encoded by SARS-CoV-2 at a glance. Med. Microbiol. Immunol. 2022, 2, 1–7. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015, 13, 307–308. [Google Scholar] [CrossRef]
- NGDC National Genomics Data Center M, Partners Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020, 48, D24–D33. [CrossRef]
- PMC-Pubmed Central. Available online: https://www.ncbi.nlm.nih.gov/pmc/ (accessed on 30 August 2022).
- Google Scholar. Available online: https://scholar.google.com/ (accessed on 30 August 2022).
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; Peddu, V.; Xie, H.; Shrestha, L.; Huang, M.-L.; Mears, M.C.; Cajimat, M.N.; Bente, D.A.; Shi, P.-Y.; Bovier, F.; et al. In vivo antiviral host response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020, 18, e3000849. [Google Scholar] [CrossRef]
- Ioannidis, I.; McNally, B.; Willette, M.; Peeples, M.E.; Chaussabel, D.; Durbin, J.E.; Ramilo, O.; Mejias, A.; Flaño, E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J. Virol. 2012, 86, 5422–5436. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Heath, E.; Jefferson, B.; Joslyn, C.; Kvinge, H.; Mitchell, H.D.; Praggastis, B.; Eisfeld, A.J.; Sims, A.C.; Thackray, L.B.; et al. Hypergraph models of biological networks to identify genes critical to pathogenic viral response. BMC Bioinform. 2021, 22, 287. [Google Scholar] [CrossRef]
- Mitchell, H.D.; Eisfeld, A.J.; Sims, A.C.; McDermott, J.E.; Matzke, M.M.; Webb-Robertson, B.-J.M.; Tilton, S.C.; Tchitchek, N.; Josset, L.; Li, C.; et al. A network integration approach to predict conserved regulators related to pathogenicity of influenza and SARS-CoV respiratory viruses. PLoS ONE 2013, 8, e69374. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Moulos, P.; Hatzis, P. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleic Acids Res. 2015, 43, e25. [Google Scholar] [CrossRef]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef]
- Bizzotto, J.; Sanchis, P.; Abbate, M.; Lage-Vickers, S.; Lavignolle, R.; Toro, A.; Olszevicki, S.; Sabater, A.; Cascardo, F.; Vazquez, E.; et al. SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. iScience 2020, 23, 101585. [Google Scholar] [CrossRef]
- Perry, M. Heatmaps: Flexible Heatmaps for Functional Genomics and Sequence Features; 2020. [Google Scholar]
- Wickham, H. ggplot2. Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots; 2020. [Google Scholar]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Segatori, V.I.; Garona, J.; Caligiuri, L.G.; Bizzotto, J.; Lavignolle, R.; Toro, A.; Sanchis, P.; Spitzer, E.; Krolewiecki, A.; Gueron, G.; et al. Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients. Viruses 2021, 13, 2084. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, A.P.; Pagotto, R.; Pórfido, J.L.; Daghero, H.; Segovia, M.; Yamasaki, K.; Varela, B.; Hill, M.; Verdes, J.M.; Duhalde Vega, M.; et al. Ivermectin reduces in vivo coronavirus infection in a mouse experimental model. Sci. Rep. 2021, 11, 7132. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Tian, L.; Luo, G.; Yu, X. Interferon-Gamma-Mediated Osteoimmunology. Front. Immunol. 2018, 9, 1508. [Google Scholar] [CrossRef]
- Körner, R.W.; Majjouti, M.; Alcazar, M.A.A.; Mahabir, E. Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2. Viruses 2020, 12, 880. [Google Scholar] [CrossRef]
- Li, S.-F.; Gong, M.-J.; Zhao, F.-R.; Shao, J.-J.; Xie, Y.-L.; Zhang, Y.-G.; Chang, H.-Y. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 51, 2377–2396. [Google Scholar] [CrossRef]
- Teijaro, J.R. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016, 16, 31–40. [Google Scholar] [CrossRef]
- Galbraith, M.D.; Kinning, K.T.; Sullivan, K.D.; Araya, P.; Smith, K.P.; Granrath, R.E.; Shaw, J.R.; Baxter, R.; Jordan, K.R.; Russell, S.; et al. Specialized interferon action in COVID-19. Proc. Natl. Acad. Sci. USA 2022, 119, e2116730119. [Google Scholar] [CrossRef]
- Bosi, E.; Bosi, C.; Rovere Querini, P.; Mancini, N.; Calori, G.; Ruggeri, A.; Canzonieri, C.; Callegaro, L.; Clementi, M.; De Cobelli, F.; et al. Interferon β-1a (IFNβ-1a) in COVID-19 patients (INTERCOP): Study protocol for a randomized controlled trial. Trials 2020, 21, 939. [Google Scholar] [CrossRef]
- Montalvo Villalba, M.C.; Valdés Ramírez, O.; Muné Jiménez, M.; Arencibia Garcia, A.; Martinez Alfonso, J.; González Baéz, G.; Roque Arrieta, R.; Rosell Simón, D.; Alvárez Gainza, D.; Sierra Vázquez, B.; et al. Interferon gamma, TGF-β1 and RANTES expression in upper airway samples from SARS-CoV-2 infected patients. Clin. Immunol. 2020, 220, 108576. [Google Scholar] [CrossRef]
- Heuberger, J.; Trimpert, J.; Vladimirova, D.; Goosmann, C.; Lin, M.; Schmuck, R.; Mollenkopf, H.-J.; Brinkmann, V.; Tacke, F.; Osterrieder, N.; et al. Epithelial response to IFN-γ promotes SARS-CoV-2 infection. EMBO Mol. Med. 2021, 13, e13191. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef] [PubMed]
- Gadotti, A.C.; de Castro Deus, M.; Telles, J.P.; Wind, R.; Goes, M.; Garcia Charello Ossoski, R.; de Padua, A.M.; de Noronha, L.; Moreno-Amaral, A.; Baena, C.P.; et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020, 289, 198171. [Google Scholar] [CrossRef]
- Bastard, P.; Gervais, A.; Le Voyer, T.; Rosain, J.; Philippot, Q.; Manry, J.; Michailidis, E.; Hoffmann, H.-H.; Eto, S.; Garcia-Prat, M.; et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 2021, 6, eabl4340. [Google Scholar] [CrossRef]
- Lee, W.-I.; Huang, J.-L.; Wu, T.-S.; Lee, M.-H.; Chen, I.-J.; Yu, K.-H.; Liu, C.-Y.; Yang, C.-H.; Hsieh, M.-Y.; Lin, Y.-L.; et al. Patients with inhibitory and neutralizing auto-antibodies to interferon-γ resemble the sporadic adult-onset phenotype of Mendelian Susceptibility to Mycobacterial Disease (MSMD) lacking Bacille Calmette-Guerin (BCG)-induced diseases. Immunobiology 2013, 218, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Liew, W.-K.; Thoon, K.-C.; Chong, C.-Y.; Tan, N.W.H.; Cheng, D.-T.; Chan, B.S.W.; Ng, M.S.Y.; Das, L.; Arkachaisri, T.; Huang, C.-H.; et al. Juvenile-Onset Immunodeficiency Secondary to Anti-Interferon-Gamma Autoantibodies. J. Clin. Immunol. 2019, 39, 512–518. [Google Scholar] [CrossRef]
- Valour, F.; Perpoint, T.; Sénéchal, A.; Kong, X.-F.; Bustamante, J.; Ferry, T.; Chidiac, C.; Ader, F. Interferon-γ Autoantibodies as Predisposing Factor for Nontuberculous Mycobacterial Infection. Emerg. Infect. Dis. 2016, 22, 1124–1126. [Google Scholar] [CrossRef]
- Halin Bergström, S.; Nilsson, M.; Adamo, H.; Thysell, E.; Jernberg, E.; Stattin, P.; Widmark, A.; Wikström, P.; Bergh, A. Extratumoral heme oxygenase-1 (HO-1) expressing macrophages likely promote primary and metastatic prostate tumor growth. PLoS ONE 2016, 11, e0157280. [Google Scholar] [CrossRef]
- Shih, H.-P.; Ding, J.-Y.; Yeh, C.-F.; Chi, C.-Y.; Ku, C.-L. Anti-interferon-γ autoantibody-associated immunodeficiency. Curr. Opin. Immunol. 2021, 72, 206–214. [Google Scholar] [CrossRef]
- Van Laarhoven, A.; Kurver, L.; Overheul, G.J.; Kooistra, E.J.; Abdo, W.F.; van Crevel, R.; Duivenvoorden, R.; Kox, M.; Ten Oever, J.; Schouten, J.; et al. Interferon gamma immunotherapy in five critically ill COVID-19 patients with impaired cellular immunity: A case series. Med. N. Y. 2021, 2, 1163–1170.e2. [Google Scholar] [CrossRef]
- Nguyen, L.S.; Ait Hamou, Z.; Gastli, N.; Chapuis, N.; Pène, F. Potential role for interferon gamma in the treatment of recurrent ventilator-acquired pneumonia in patients with COVID-19: A hypothesis. Intensive Care Med. 2021, 47, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, T.; Kubli, S.P.; Yoshinaga, S.K.; Pfeffer, K.; Mak, T.W. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020, 27, 3209–3225. [Google Scholar] [CrossRef] [PubMed]
- NIH Kinase Inhibitors. COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/therapies/immunomodulators/kinase-inhibitors/ (accessed on 30 August 2022).
- Ely, E.W.; Ramanan, A.V.; Kartman, C.E.; de Bono, S.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Saraiva, J.F.K.; Chakladar, S.; Marconi, V.C.; et al. Efficacy and safety of baricitinib plus standard of care for the treatment of critically ill hospitalised adults with COVID-19 on invasive mechanical ventilation or extracorporeal membrane oxygenation: An exploratory, randomised, placebo-controlled trial. Lancet. Respir. Med. 2022, 10, 327–336. [Google Scholar] [CrossRef]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; de Cassia Pellegrini, R.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet. Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- RECOVERY Collaborative Group Baricitinib in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial and updated meta-analysis. Lancet 2022, 400, 359–368. [CrossRef]
- Garcia-Del-Barco, D.; Risco-Acevedo, D.; Berlanga-Acosta, J.; Martos-Benítez, F.D.; Guillén-Nieto, G. Revisiting Pleiotropic Effects of Type I Interferons: Rationale for Its Prophylactic and Therapeutic Use Against SARS-CoV-2. Front. Immunol. 2021, 12, 655528. [Google Scholar] [CrossRef]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551–566. [Google Scholar] [CrossRef]
- Visan, I. The interferon signature. Nat. Immunol. 2017, 18, 151. [Google Scholar] [CrossRef]
- Bińkowski, J.; Taryma-Leśniak, O.; Łuczkowska, K.; Niedzwiedź, A.; Lechowicz, K.; Strapagiel, D.; Jarczak, J.; Davalos, V.; Pujol, A.; Esteller, M.; et al. Epigenetic activation of antiviral sensors and effectors of interferon response pathways during SARS-CoV-2 infection. Biomed. Pharmacother. 2022, 153, 113396. [Google Scholar] [CrossRef]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Zhao, C.; Collins, M.N.; Hsiang, T.-Y.; Krug, R.M. Interferon-induced ISG15 pathway: An ongoing virus-host battle. Trends Microbiol. 2013, 21, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, J.; Yan, H.; Huang, J.; Wang, F.; Liu, T.; Zeng, L.; Zhou, F. ISGylation in Innate Antiviral Immunity and Pathogen Defense Responses: A Review. Front. Cell Dev. Biol. 2021, 9, 788410. [Google Scholar] [CrossRef] [PubMed]
- Vere, G.; Alam, M.R.; Farrar, S.; Kealy, R.; Kessler, B.M.; O’Brien, D.P.; Pinto-Fernández, A. Targeting the Ubiquitylation and ISGylation Machinery for the Treatment of COVID-19. Biomolecules 2022, 12, 300. [Google Scholar] [CrossRef]
- Schwartz, S.L.; Park, E.N.; Vachon, V.K.; Danzy, S.; Lowen, A.C.; Conn, G.L. Human OAS1 activation is highly dependent on both RNA sequence and context of activating RNA motifs. Nucleic Acids Res. 2020, 48, 7520–7531. [Google Scholar] [CrossRef] [PubMed]
- Danziger, O.; Patel, R.S.; DeGrace, E.J.; Rosen, M.R.; Rosenberg, B.R. Inducible CRISPR activation screen for interferon-stimulated genes identifies OAS1 as a SARS-CoV-2 restriction factor. PLoS Pathog. 2022, 18, e1010464. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.; Butler-Laporte, G.; Khan, A.; Drivas, T.G.; Peloso, G.M.; Nakanishi, T.; Verma, A.; Kiryluk, K.; Richards, J.B.; Zeberg, H. Alternative splicing of OAS1 alters the risk for severe COVID-19. medRxiv 2021, 25. [Google Scholar] [CrossRef]
- Huffman, J.E.; Butler-Laporte, G.; Khan, A.; Pairo-Castineira, E.; Drivas, T.G.; Peloso, G.M.; Nakanishi, T.; Ganna, A.; Verma, A.; Baillie, J.K.; et al. Multi-ancestry fine mapping implicates OAS1 splicing in risk of severe COVID-19. Nat. Genet. 2022, 54, 125–127. [Google Scholar] [CrossRef]
- Banday, A.R.; Stanifer, M.L.; Florez-Vargas, O.; Onabajo, O.O.; Papenberg, B.W.; Zahoor, M.A.; Mirabello, L.; Ring, T.J.; Lee, C.-H.; Albert, P.S.; et al. Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat. Genet. 2022, 54, 1103–1116. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A prenylated dsRNA sensor protects against severe COVID-19. Science 2021, 374, eabj3624. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Zhou, S.; Butler-Laporte, G.; Nakanishi, T.; Morrison, D.R.; Afilalo, J.; Afilalo, M.; Laurent, L.; Pietzner, M.; Kerrison, N.; Zhao, K.; et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med. 2021, 27, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Hurgin, V.; Novick, D.; Werman, A.; Dinarello, C.A.; Rubinstein, M. Antiviral and immunoregulatory activities of IFN-gamma depend on constitutively expressed IL-1alpha. Proc. Natl. Acad. Sci. USA 2007, 104, 5044–5049. [Google Scholar] [CrossRef] [PubMed]
- Van de Veerdonk, F.L.; Giamarellos-Bourboulis, E.; Pickkers, P.; Derde, L.; Leavis, H.; van Crevel, R.; Engel, J.J.; Wiersinga, W.J.; Vlaar, A.P.J.; Shankar-Hari, M.; et al. A guide to immunotherapy for COVID-19. Nat. Med. 2022, 28, 39–50. [Google Scholar] [CrossRef]
- McKimm-Breschkin, J.L. Management of influenza virus infections with neuraminidase inhibitors: Detection, incidence, and implications of drug resistance. Treat. Respir. Med. 2005, 4, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S. The role of antivirals in the control of influenza. Vaccine 2003, 21, 1796–1800. [Google Scholar] [CrossRef]

| Gene | Forward (5′-3′) | Reverse (5′-3′) | T ann (°C) * |
|---|---|---|---|
| MX1 | AGGACCATCGGAATCTTGAC | TCAGGTGGAACACGAGGTTC | 60 |
| MX2 | GGCAGAGGCAACCAAGAAAGA | AACGGGAGCGATTTTTGGA | 60 |
| ISG15 | GTCCTGCTGGTGGTGGACAAA | GTCCTGCTGGTGGTGGACAAA | 61 |
| OAS1 | GGGATTTCGGACGGTCTTGG | TCTCCACCACCCAAGTTTCC | 60 |
| PPIA | GGTATAAAAGGGGCGGGAGG | CTGCAAACAGCTCAAAGGAGAC | 60 |
| Mx1 | TGCCAGGACCAGGTTTACAAG | CCCCTTTTGAGGAAACTGAGA | 58 |
| Mx2 | CCTATTCACCAGGCTCCGAA | TCTCGTCCACGGTACTGCTT | 58 |
| Isg15 | TGAGAGCAAGCAGCCAGAAG | CCCCCAGCATCTTCACCTTT | 57 |
| Oas1 | ACTTCCTGAACTGTCGCCC | ACTCGACTCCCATACTCCCAG | 61 |
| Gapdh | TGCCAAGGCTGTGGGCAAGG | CGAAGGTGGAAGAGTGGG | 60 |
| MHV | GGAACTTCTCGTTGGGCATTATACT | ACCACAAGATTATCATTTTCACAACATA | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toro, A.; Lage-Vickers, S.; Bizzotto, J.; Vilicich, F.; Sabater, A.; Pascual, G.; Ledesma-Bazan, S.; Sanchis, P.; Ruiz, M.S.; Arevalo, A.P.; et al. Pin-Pointing the Key Hubs in the IFN-γ Pathway Responding to SARS-CoV-2 Infection. Viruses 2022, 14, 2180. https://doi.org/10.3390/v14102180
Toro A, Lage-Vickers S, Bizzotto J, Vilicich F, Sabater A, Pascual G, Ledesma-Bazan S, Sanchis P, Ruiz MS, Arevalo AP, et al. Pin-Pointing the Key Hubs in the IFN-γ Pathway Responding to SARS-CoV-2 Infection. Viruses. 2022; 14(10):2180. https://doi.org/10.3390/v14102180
Chicago/Turabian StyleToro, Ayelen, Sofia Lage-Vickers, Juan Bizzotto, Felipe Vilicich, Agustina Sabater, Gaston Pascual, Sabrina Ledesma-Bazan, Pablo Sanchis, Maria Sol Ruiz, Ana Paula Arevalo, and et al. 2022. "Pin-Pointing the Key Hubs in the IFN-γ Pathway Responding to SARS-CoV-2 Infection" Viruses 14, no. 10: 2180. https://doi.org/10.3390/v14102180
APA StyleToro, A., Lage-Vickers, S., Bizzotto, J., Vilicich, F., Sabater, A., Pascual, G., Ledesma-Bazan, S., Sanchis, P., Ruiz, M. S., Arevalo, A. P., Porfido, J. L., Abbate, M., Seniuk, R., Labanca, E., Anselmino, N., Navone, N. M., Alonso, D. F., Vazquez, E., Crispo, M., ... Gueron, G. (2022). Pin-Pointing the Key Hubs in the IFN-γ Pathway Responding to SARS-CoV-2 Infection. Viruses, 14(10), 2180. https://doi.org/10.3390/v14102180










