Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations
Abstract
1. Introduction
2. Materials and Methods
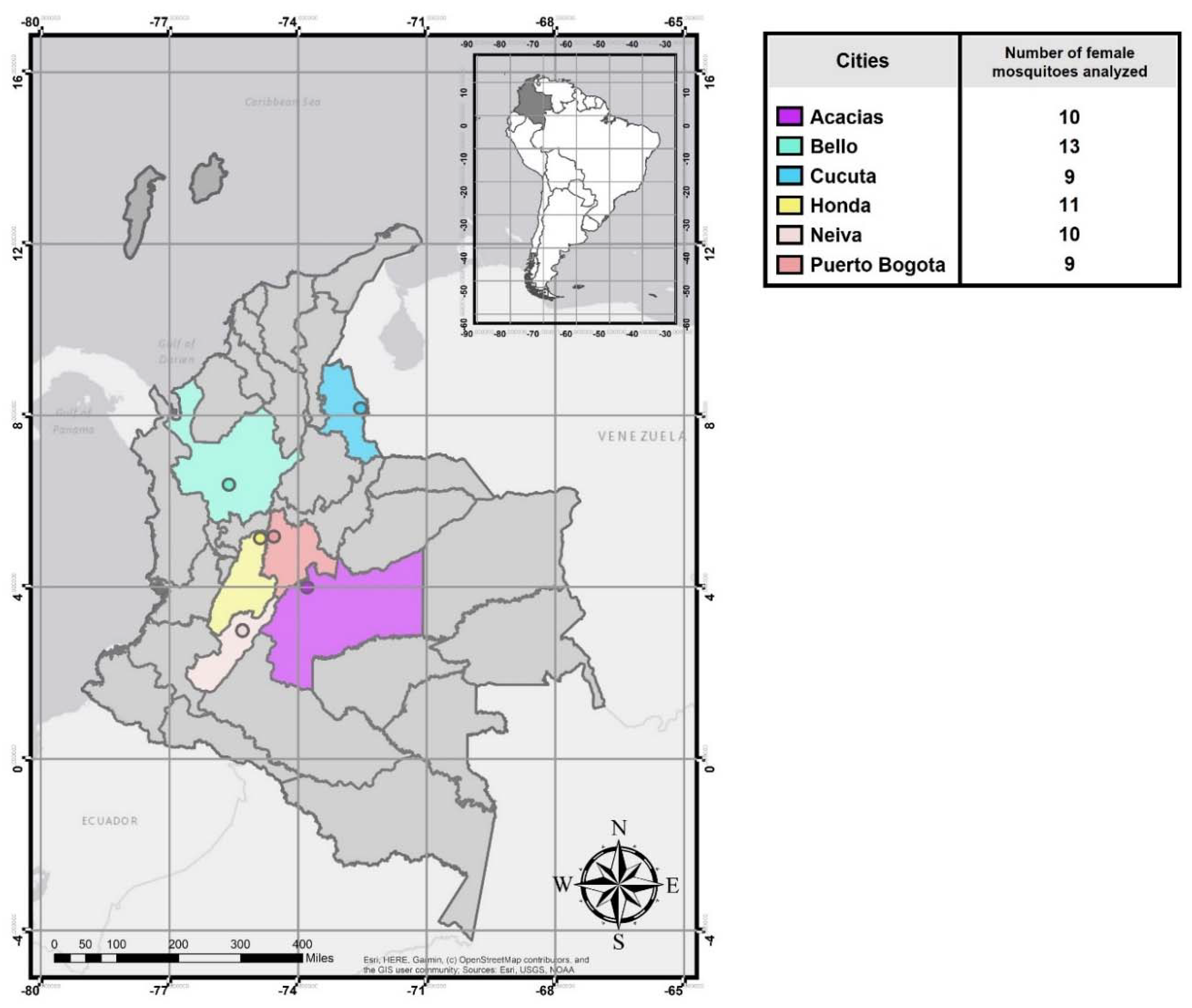
2.1. Mosquito Collection
2.2. ZIKV detection in Field Females
2.3. Determination of lambda–cyhalothrin Resistance Profile and Allele-Specific PCR (AS-PCR) for the kdr Mutation V1016I
2.4. Preparation of Genomic DNA and Library for Metagenome 16S Sequencing
2.5. Microbial Community Analysis
2.6. Prediction of Functional Genes
3. Results
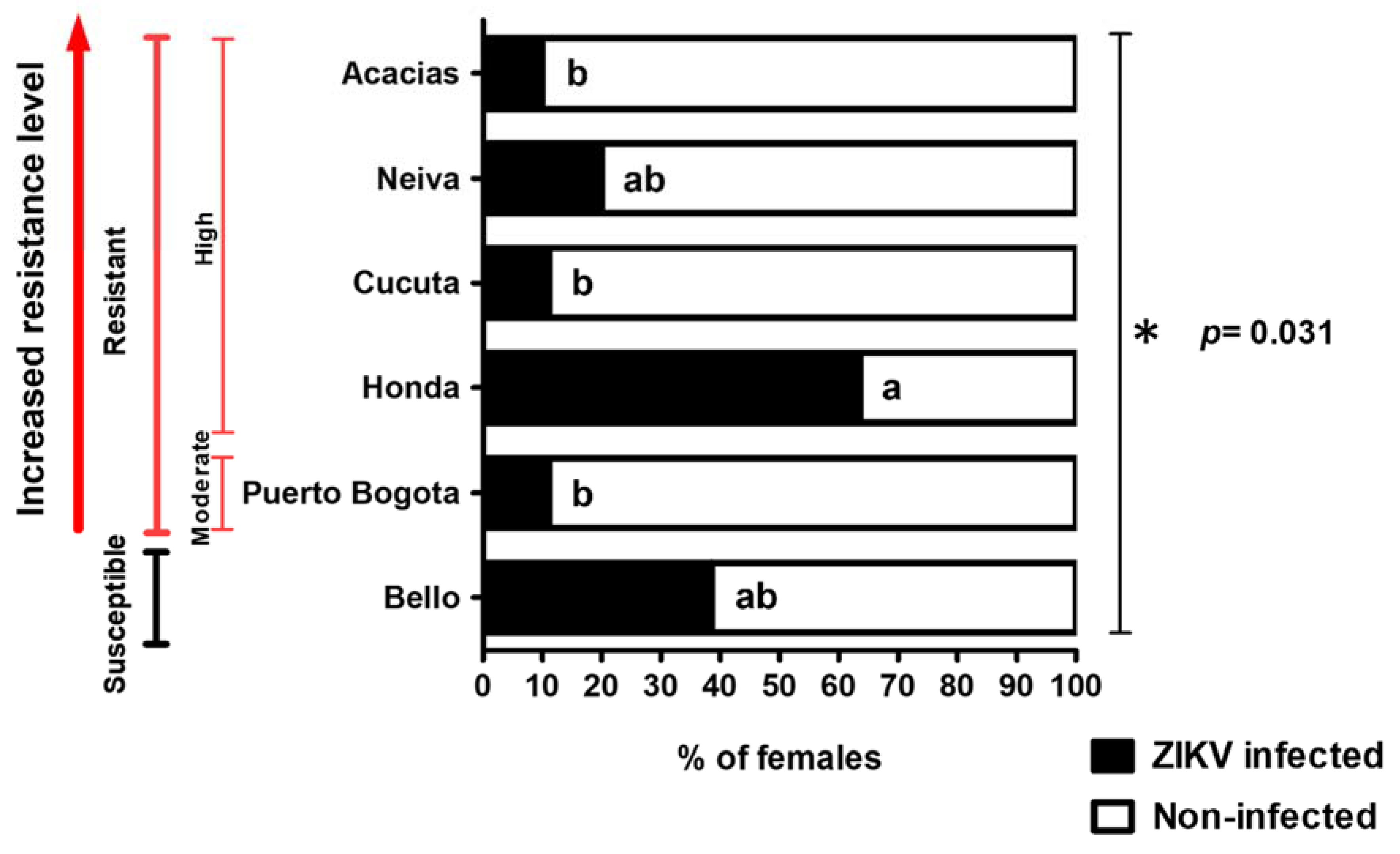
3.1. Natural ZIKV Infection in Colombian Ae. aegypti Females
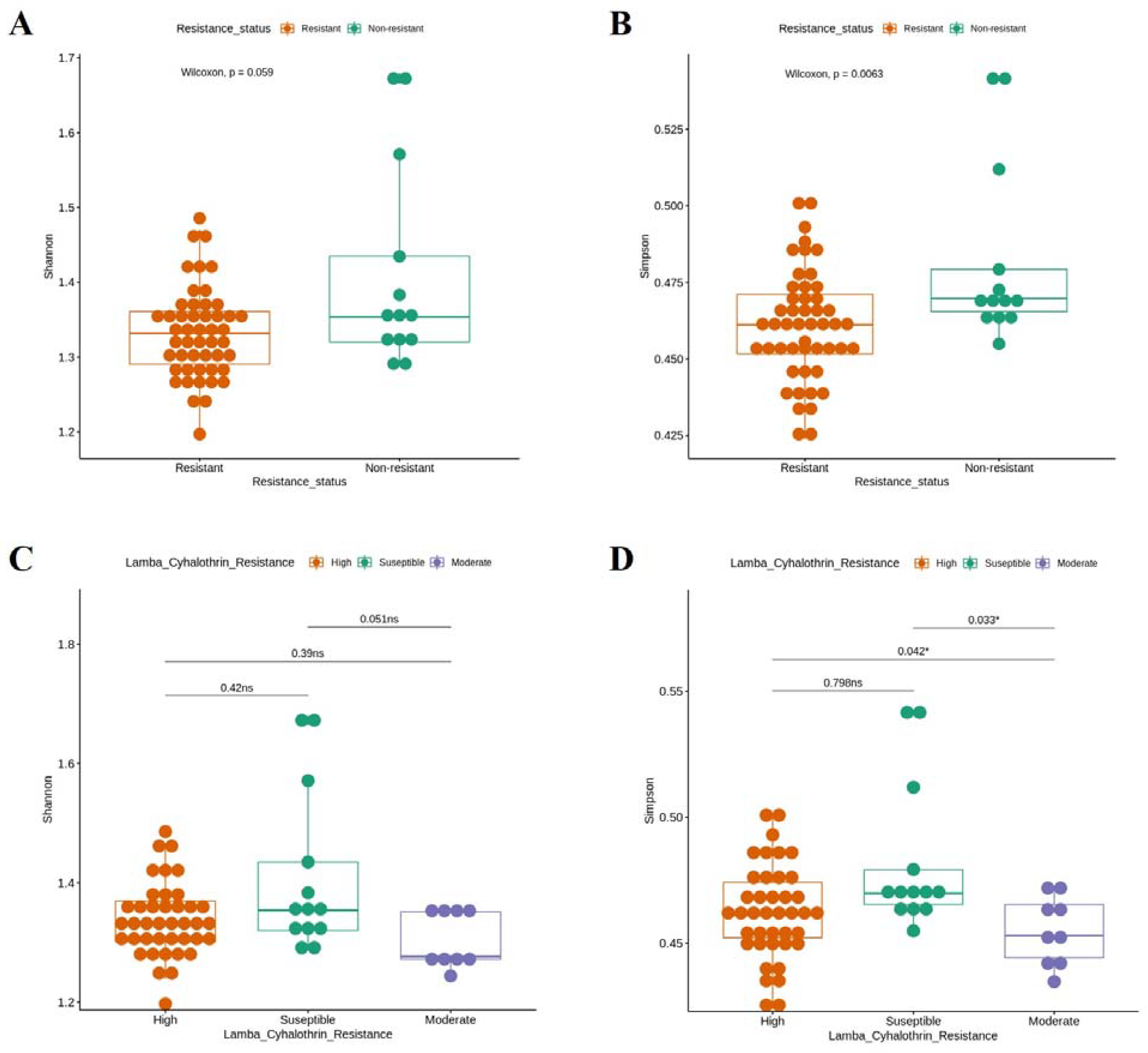
3.2. Bacterial Diversity in Colombian Ae. aegypti Populations According to Natural ZIKV Infection and lambda–cyhalothrin Resistance Status
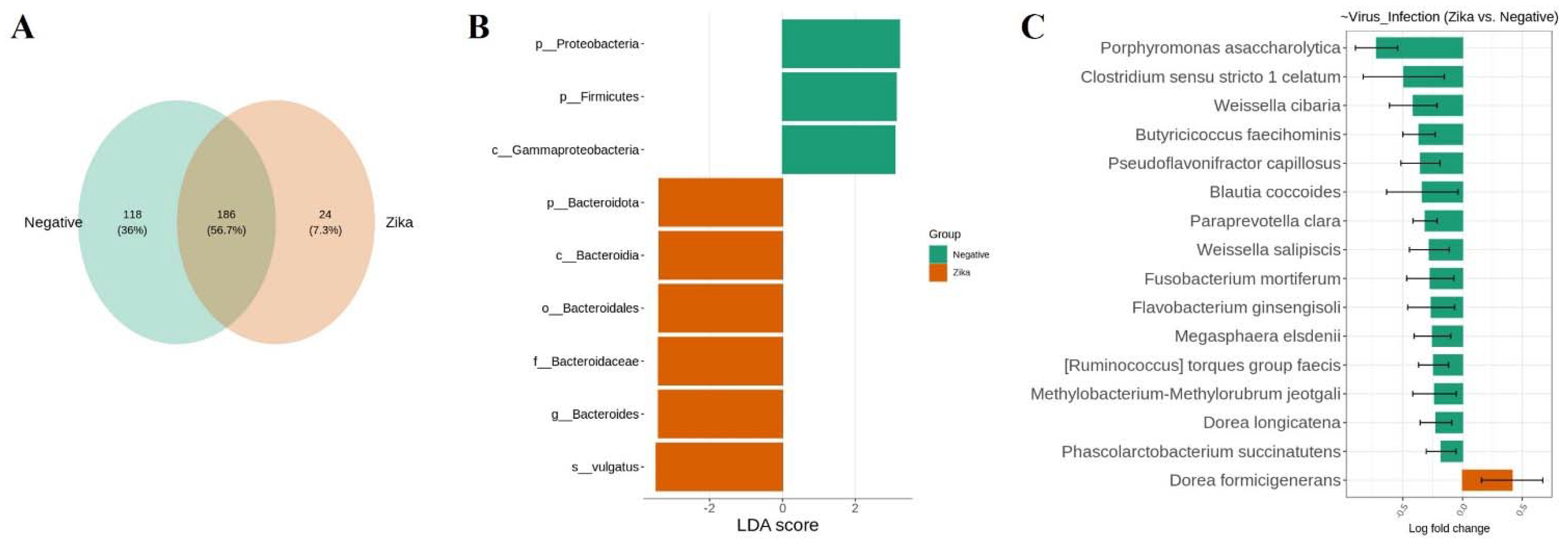
3.3. The Bacterial Signature Associated with Natural ZIKV Infection in Colombian Ae. Aegypti Midguts
3.4. Bacterial Signature in Ae. aegypti Midguts Associated with lambda–cyhalothrin Susceptibility and High and Moderate Resistance
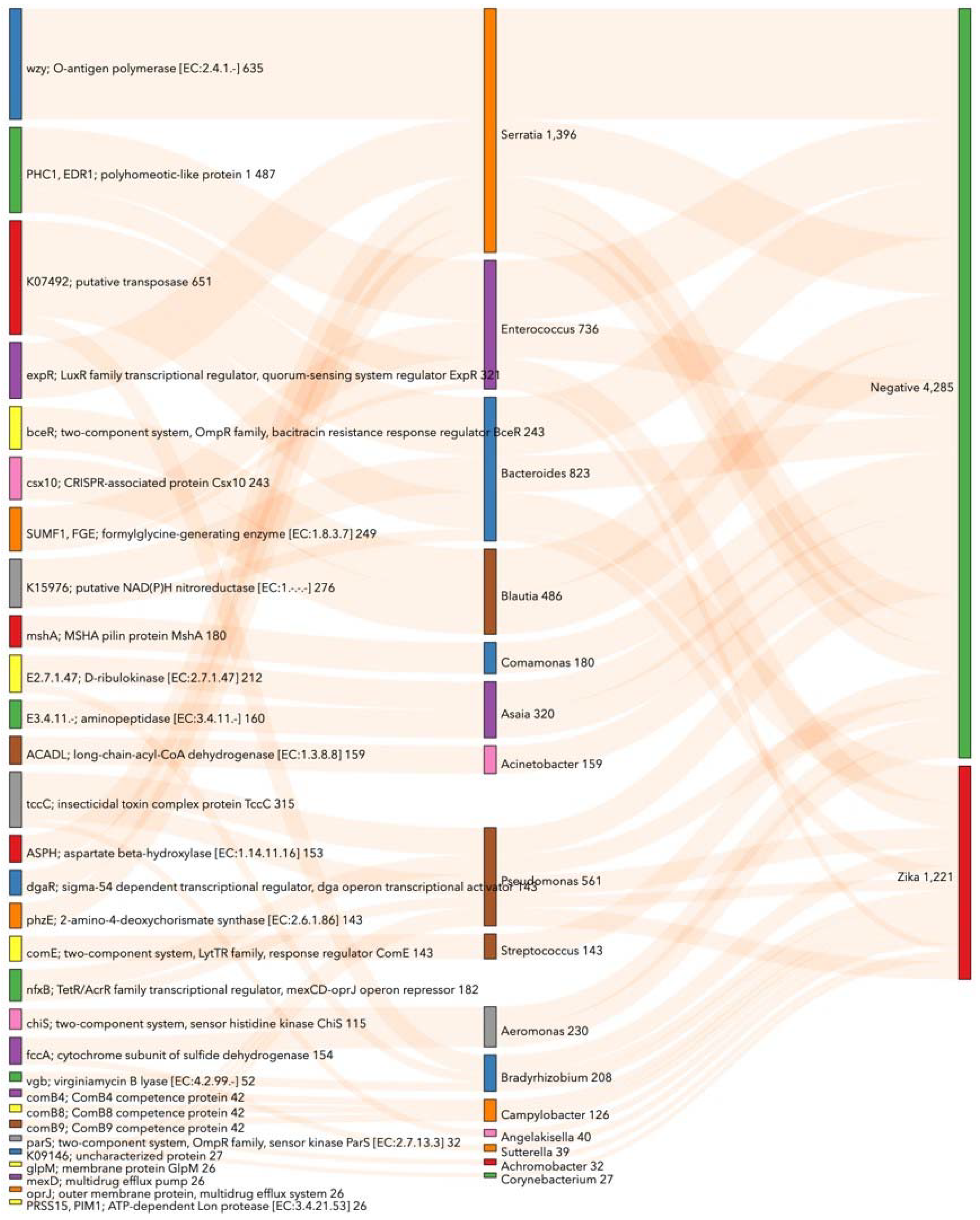
3.5. Prediction of Functional Metabolic Profiling on Bacterial KEGG Genes Associated with Ae. aegypti Midgut Naturally Infected with ZIKV
3.6. Prediction of Functional Metabolic Profiling on Bacterial KEGG Genes Associated with lambda–cyhalothrin Susceptibility and Resistance in Ae. aegypti
4. Discussion
4.1. Evidence of Natural ZIKV Infection in Rural and Urban Ae. aegypti Populations in Colombia with Different Degrees of Resistance to lambda–cyhalothrin
4.2. Bacterial Signature in Ae. aegypti Midgut Related to Natural ZIKV Infection and Resistance to lambda–cyhalothrin
4.3. Predicted Bacterial KEGG Pathways Related to Natural ZIKV Infection and Insecticide Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carvalho, V.L.; Long, M.T. Perspectives on New Vaccines against Arboviruses Using Insect-Specific Viruses as Platforms. Vaccines 2021, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Santacoloma Varón, L.; Chaves Córdoba, B.; Brochero, H.L. Susceptibilidad de Aedes aegypti a DDT, deltametrina y lambdacialotrina en Colombia. Rev. Panam. Salud. Publica 2010, 27, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Serrano, R.; Gomez-Camargo, D.; Ponce-Garcia, G.; Flores, A.E. Susceptibility to insecticides and resistance mechanisms in Aedes aegypti from the Colombian Caribbean Region. Pestic. Biochem. Physiol. 2014, 116, 63–73. [Google Scholar] [CrossRef]
- Granada, Y.; Mejía-Jaramillo, A.; Strode, C.; Triana-Chavez, O. A Point Mutation V419L in the Sodium Channel Gene from Natural Populations of Aedes aegypti Is Involved in Resistance to λ-Cyhalothrin in Colombia. Insects 2018, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Aponte, A.; Penilla, R.P.; Rodríguez, A.D.; Ocampo, C.B. Mechanisms of pyrethroid resistance in Aedes (Stegomyia) aegypti from Colombia. Acta Trop. 2019, 191, 146–154. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, K.; Chinazzi, M.; Pastore y Piontti, A.; Dean, N.E.; Rojas, D.P.; Merler, S.; Mistry, D.; Poletti, P.; Rossi, L.; et al. Spread of Zika virus in the Americas. Proc. Natl. Acad. Sci. USA 2017, 114, E4334–E4343. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.C.; Calisher, C.H. Emergence of Human Arboviral Diseases in the Americas, 2000–2016. Vector-Borne Zoonotic Dis. 2016, 16, 295–301. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, T.; Lai, Z.; Zhang, Z.; Jia, Z.; Zhou, G.; Williams, T.; Xu, J.; Gu, J.; Zhou, X.; et al. Competence of Aedes aegypti, Ae. albopictus, and Culex quinquefasciatus Mosquitoes as Zika Virus Vectors, China. Emerg. Infect. Dis. 2017, 23, 1085–1091. [Google Scholar] [CrossRef]
- Vorou, R. Zika virus, vectors, reservoirs, amplifying hosts, and their potential to spread worldwide: What we know and what we should investigate urgently. Int. J. Infect. Dis. 2016, 48, 85–90. [Google Scholar] [CrossRef]
- Pérez-Pérez, J.; Rojo-Ospina, R.A.; Henao, E.; García-Huertas, P.; Triana-Chavez, O.; Rúa-Uribe, G. Natural infection of Aedes aegypti, Ae. albopictus and Culex spp. with Zika virus in Medellin, Colombia. CES Med. 2019, 33, 175–181. [Google Scholar] [CrossRef]
- Boeuf, P.; Drummer, H.E.; Richards, J.S.; Scoullar, M.J.L.; Beeson, J.G. The global threat of Zika virus to pregnancy: Epidemiology, clinical perspectives, mechanisms, and impact. BMC Med. 2016, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Morales, A.J.; Willamil-Gómez, W.E. El reto de Zika en Colombia y América Latina: Una urgencia sanitaria internacional. Infectio 2016, 20, 59–61. [Google Scholar] [CrossRef]
- Perez, F.; Llau, A.; Gutierrez, G.; Bezerra, H.; Coelho, G.; Ault, S.; Brandao Barbiratto, S.; Carballo de Resende, M. The decline of dengue in the Americas in 2017: Discussion of multiple hypotheses. Trop. Med. Int. Health 2019, 24, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Brito, A.F.; Machado, L.C.; Siconelli, M.J.L.; Oidtman, R.J.; Fauver, J.R.; de Oliveira Carvalho, R.D.; Dezordi, F.Z.; Pereira, M.R.; de Castro-Jorge, L.A.; Minto, E.C.M.; et al. Lying in wait: The resurgence of dengue virus after the Zika epidemic in Brazil. Epidemiology 2020. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 10 de 2016. BES-Sivigila. 2016. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2016%20Bolet%C3%ADn%20epidemiol%C3%B3gico%20semana%2010.pdf (accessed on 21 October 2021).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana EPIDEMIOLÓGICA número 52 de 2016. BES-Sivigila. 2016. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2016%20Bolet%C3%ADn%20epidemiol%C3%B3gico%20semana%2052%20-.pdf (accessed on 21 October 2021).
- Instituto Nacional de Salud. Informe del Evento Enfermedad por Virus Zika Colombia, 2017. 2018. Available online: https://www.ins.gov.co/buscador-eventos/Informesdeevento/ZIKA%202017.pdf (accessed on 21 October 2021).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 52 de 2018. BES-Sivigila. 2018. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2018%20Bolet%C3%ADn%20epidemiol%C3%B3gico%20semana%2052.pdf (accessed on 21 October 2021).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 52 de 2019. BES-Sivigila. 2019. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2019_Boletin_epidemiologico_semana_52.pdf (accessed on 21 October 2021).
- Guégan, M.; Zouache, K.; Démichel, C.; Minard, G.; Tran Van, V.; Potier, P.; Mavingui, P.; Valiente Moro, C. The mosquito holobiont: Fresh insight into mosquito-microbiota interactions. Microbiome 2018, 6, 49. [Google Scholar] [CrossRef]
- Joanne, S.; Vythilingam, I.; Yugavathy, N.; Leong, C.S.; Wong, M.L.; AbuBakar, S. Distribution and dynamics of Wolbachia infection in Malaysian Aedes albopictus. Acta Trop. 2015, 148, 38–45. [Google Scholar] [CrossRef]
- Ye, Y.H.; Carrasco, A.M.; Frentiu, F.D.; Chenoweth, S.F.; Beebe, N.W.; van den Hurk, A.F.; Simmons, C.P.; O’Neill, S.L.; McGraw, E.A. Wolbachia Reduces the Transmission Potential of Dengue-Infected Aedes aegypti. PLoS Negl. Trop. Dis. 2015, 9, e0003894. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Souza-Neto, J.; Torres Cosme, R.; Rovira, J.; Ortiz, A.; Pascale, J.M.; Dimopoulos, G.; O’Neill, S.L. Reciprocal Tripartite Interactions between the Aedes aegypti Midgut Microbiota, Innate Immune System and Dengue Virus Influences Vector Competence. PLoS Negl. Trop Dis. 2012, 6, e1561. [Google Scholar] [CrossRef]
- Wu, P.; Sun, P.; Nie, K.; Zhu, Y.; Shi, M.; Xiao, C.; Liu, H.; Liu, Q.; Zhao, T.; Chen, X.; et al. A Gut Commensal Bacterium Promotes Mosquito Permissiveness to Arboviruses. Cell Host Microbe 2019, 25, 101–112.e5. [Google Scholar] [CrossRef]
- Villegas, L.E.M.; Campolina, T.B.; Barnabe, N.R.; Orfano, A.S.; Chaves, B.A.; Norris, D.E.; Pimenta, P.F.P.; Secundino, N.F.C. Zika virus infection modulates the bacterial diversity associated with Aedes aegypti as revealed by metagenomic analysis. PLoS ONE 2018, 13, e0190352. [Google Scholar] [CrossRef]
- Zink, S.; Van Slyke, G.; Palumbo, M.; Kramer, L.; Ciota, A. Exposure to West Nile Virus Increases Bacterial Diversity and Immune Gene Expression in Culex pipiens. Viruses 2015, 7, 5619–5631. [Google Scholar] [CrossRef] [PubMed]
- Zouache, K.; Michelland, R.J.; Failloux, A.B.; Grundmann, G.L.; Mavingui, P. Chikungunya virus impacts the diversity of symbiotic bacteria in mosquito vector. Mol. Ecol. 2012, 21, 2297–2309. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Sun, P.; Yu, X.; Wang, P.; Cheng, G. Roles of Symbiotic Microorganisms in Arboviral Infection of Arthropod Vectors. Trends Parasitol. 2020, 36, 607–615. [Google Scholar] [CrossRef]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia Symbiont in Aedes aegypti Limits Infection with Dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Short, S.M.; Bahia, A.C.; Saraiva, R.G.; Dong, Y.; Kang, S.; Tripathi, A.; Mlambo, G.; Dimopoulos, G. Chromobacterium Csp_P Reduces Malaria and Dengue Infection in Vector Mosquitoes and Has Entomopathogenic and In Vitro Anti-pathogen Activities. PLoS Pathog. 2014, 10, e1004398. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The Endosymbiotic Bacterium Wolbachia Induces Resistance to Dengue Virus in Aedes aegypti. PLoS Pathog. 2010, 6, e1000833. [Google Scholar] [CrossRef]
- Joubert, D.A.; Walker, T.; Carrington, L.B.; De Bruyne, J.T.; Kien, D.H.T.; Hoang, N.L.T.; Chau, N.V.V.; Iturbe-Ormaetxe, I.; Simmons, C.P.; O’Neill, S.L. Establishment of a Wolbachia Superinfection in Aedes aegypti Mosquitoes as a Potential Approach for Future Resistance Management. PLoS Pathog. 2016, 12, e1005434. [Google Scholar] [CrossRef]
- Pan, X.; Zhou, G.; Wu, J.; Bian, G.; Lu, P.; Raikhel, A.S.; Xi, Z. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2012, 109, E23–E31. [Google Scholar] [CrossRef]
- Chouin-Carneiro, T.; Ant, T.H.; Herd, C.; Louis, F.; Failloux, A.B.; Sinkins, S.P. Wolbachia strain wAlbA blocks Zika virus transmission in Aedes aegypti. Med. Vet. Entomol. 2020, 34, 116–119. [Google Scholar] [CrossRef]
- Pan, X.; Pike, A.; Joshi, D.; Bian, G.; McFadden, M.J.; Lu, P.; Liang, X.; Zhang, F.; Raikhel, A.S.; Xi, Z. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 2018, 12, 277–288. [Google Scholar] [CrossRef]
- Dutra, H.L.C.; Rocha, M.N.; Dias, F.B.S.; Mansur, S.B.; Caragata, E.P.; Moreira, L.A. Wolbachia Blocks Currently Circulating Zika Virus Isolates in Brazilian Aedes aegypti Mosquitoes. Cell Host Microbe 2016, 19, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Frentiu, F.D.; Moreira, L.A.; O’Neill, S.L.; Asgari, S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. USA 2011, 108, 9250–9255. [Google Scholar] [CrossRef] [PubMed]
- Kambris, Z.; Blagborough, A.M.; Pinto, S.B.; Blagrove, M.S.C.; Godfray, H.C.J.; Sinden, R.E.; Sinkins, S.P. Wolbachia Stimulates Immune Gene Expression and Inhibits Plasmodium Development in Anopheles gambiae. PLoS Pathog. 2010, 6, e1001143. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Joshi, D.; Dong, Y.; Lu, P.; Zhou, G.; Pan, X.; Xu, Y.; Dimopoulos, G.; Xi, Z. Wolbachia Invades Anopheles stephensi Populations and Induces Refractoriness to Plasmodium Infection. Science 2013, 340, 748–751. [Google Scholar] [CrossRef]
- Apte-Deshpande, A.; Paingankar, M.; Gokhale, M.D.; Deobagkar, D.N. Serratia odorifera a Midgut Inhabitant of Aedes aegypti Mosquito Enhances Its Susceptibility to Dengue-2 Virus. PLoS ONE 2012, 7, e40401. [Google Scholar] [CrossRef]
- Apte-Deshpande, A.; Paingankar, M.; Gokhale, M.D.; Deobagkar, D.N. Serratia odorifera mediated enhancement in susceptibility of Aedes aegypti for chikungunya virus. Indian J. Med. Res. 2014, 139, 762–768. [Google Scholar]
- Rio, R.V.M.; Jozwick, A.K.S.; Savage, A.F.; Sabet, A.; Vigneron, A.; Wu, Y.; Aksoy, S.; Weiss, B.L. Mutualist-Provisioned Resources Impact Vector Competency. mBio 2019, 10, e00018-19. [Google Scholar] [CrossRef]
- Coon, K.L.; Brown, M.R.; Strand, M.R. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 2016, 25, 5806–5826. [Google Scholar] [CrossRef]
- Wang, Y.; Gilbreath, T.M.; Kukutla, P.; Yan, G.; Xu, J. Dynamic Gut Microbiome across Life History of the Malaria Mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar]
- Oliveira, J.H.M.; Gonçalves, R.L.S.; Lara, F.A.; Dias, F.A.; Gandara, A.C.P.; Menna-Barreto, R.F.S.; Edwards, M.C.; Laurindo, F.R.M.; Silva-Neto, M.A.C.; Sorgine, M.H.F.; et al. Blood Meal-Derived Heme Decreases ROS Levels in the Midgut of Aedes aegypti and Allows Proliferation of Intestinal Microbiota. PLoS Pathog. 2011, 7, e1001320. [Google Scholar] [CrossRef]
- Beckmann, J.F.; Ronau, J.A.; Hochstrasser, M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2017, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Cortés, A.; Mejia-Jaramillo, A.M.; Granada, Y.; Coatsworth, H.; Lowenberger, C.; Triana-Chavez, O. The Midgut Microbiota of Colombian Aedes aegypti Populations with Different Levels of Resistance to the Insecticide lambda-cyhalothrin. Insects 2020, 11, 584. [Google Scholar] [CrossRef] [PubMed]
- Scates, S.S.; O’Neal, S.T.; Anderson, T.D. Bacteria-mediated modification of insecticide toxicity in the yellow fever mosquito, Aedes aegypti. Pestic. Biochem. Physiol. 2019, 161, 77–85. [Google Scholar] [CrossRef]
- Muturi, E.J.; Dunlap, C.; Smartt, C.T.; Shin, D. Resistance to permethrin alters the gut microbiota of Aedes aegypti. Sci. Rep. 2021, 11, 14406. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Shen, R.X.; Xing, D.; Zhao, C.P.; Gao, H.T.; Wu, J.H.; Zhang, N.; Zhang, H.D.; Chen, Y.; Zhao, T.Y.; et al. Metagenome Sequencing Reveals the Midgut Microbiota Makeup of Culex pipiens quinquefasciatus and Its Possible Relationship with Insecticide Resistance. Front. Microbiol. 2021, 12, 625539. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2021.625539 (accessed on 18 July 2022). [CrossRef] [PubMed]
- De Salud, I.N. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 13 de 2020. BES-Sivigila. 2020. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2020_Boletin_epidemiologico_semana_13.pdf (accessed on 24 April 2022).
- Padilla, J.C.; Rojas, D.P.; Sáenz Gómez, R. Dengue en Colombia: Epidemiología de la Reemergencia a la Hiperendemia, 1st ed.; Guías de Impresión LTDA.: Bogotá, Colombia, 2012. [Google Scholar]
- Gutierrez-Barbosa, H.; Medina-Moreno, S.; Zapata, J.C.; Chua, J.V. Dengue Infections in Colombia: Epidemiological Trends of a Hyperendemic Country. Trop. Med. Infect. Dis. 2020, 5, 156. [Google Scholar] [CrossRef]
- Caicedo, P.A.; Barón, O.L.; Pérez, M.; Alexander, N.; Lowenberger, C.; Ocampo, C.B. Selection of Aedes aegypti (Diptera: Culicidae) strains that are susceptible or refractory to Dengue-2 virus. Can. Entomol. 2013, 145, 273–282. [Google Scholar] [CrossRef]
- Serrato, I.M.; Caicedo, P.A.; Orobio, Y.; Lowenberger, C.; Ocampo, C.B. Vector competence and innate immune responses to dengue virus infection in selected laboratory and field-collected Stegomyia aegypti (=Aedes aegypti). Med. Vet. Entomol. 2017, 31, 312–319. [Google Scholar] [CrossRef]
- Arévalo-Cortés, A.; Granada, Y.; Torres, D.; Triana-Chavez, O. Differential Hatching, Development, Oviposition, and Longevity Patterns among Colombian Aedes aegypti Populations. Insects 2022, 13, 536. [Google Scholar] [CrossRef] [PubMed]
- Rueda, L.M. Pictorial Keys for the Identification of Mosquitoes (Diptera:Culicidae) Associated with Dengue Virus Transmission; Magnolia Press: Auckland, New Zealand, 2004; pp. 1–60. [Google Scholar]
- Balm, M.N.D.; Lee, C.K.; Lee, H.K.; Chiu, L.; Koay, E.S.C.; Tang, J.W. A diagnostic polymerase chain reaction assay for Zika virus. J. Med. Virol. 2012, 84, 1501–1505. [Google Scholar] [CrossRef]
- WHO. Guidelines for Laboratory and Field Testing of Mosquito Larvicides. 2005. Available online: https://apps.who.int/iris/bitstream/handle/10665/69101/WHO_CDS_WHOPES_GCDPP_2005.13.pdf?sequence=1&isAllowed=y (accessed on 25 August 2020).
- Li, C.X.; Kaufman, P.E.; Xue, R.D.; Zhao, M.H.; Wang, G.; Yan, T.; Guo, X.X.; Zhang, Y.M.; Dong, Y.D.; Xing, D.; et al. Relationship between insecticide resistance and kdr mutations in the dengue vector Aedes aegypti in Southern China. Parasites Vectors 2015, 8, 325. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.5–7. Available online: https://github.com/vegandevs/vegan (accessed on 28 November 2020).
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Pérez-Castro, R.; Castellanos, J.E.; Olano, V.A.; Matiz, M.I.; Jaramillo, J.F.; Vargas, S.L.; Sarmiento, D.M.; Stenström, T.A.; Overgaard, H.J. Detection of all four dengue serotypes in Aedes aegypti female mosquitoes collected in a rural area in Colombia. Mem. Inst. Oswaldo Cruz 2016, 111, 233–240. [Google Scholar] [CrossRef]
- Olano, V.A. Aedes aegypti en el área rural: Implicaciones en salud pública. Biomédica 2016, 36, 169–173. [Google Scholar] [CrossRef]
- Olano, V.A.; Matiz, M.I.; Lenhart, A.; Cabezas, L.; Vargas, S.L.; Jaramillo, J.F.; Sarmiento, D.; Alexander, N.; Stenström, T.A.; Overgaard, H.J. Schools as Potential Risk Sites for Vector-Borne Disease Transmission: Mosquito Vectors in Rural Schools in Two Municipalities in Colombia. J. Am. Mosq. Control. Assoc. 2015, 31, 212–222. [Google Scholar] [CrossRef]
- DANE. Censo Nacional de Población y Vivienda 2018—Colombia. Geoportal. 2018. Available online: https://geoportal.dane.gov.co/geovisores/sociedad/cnpv-2018/?lt=5.179007980008946&lg=-74.8001416124999&z=11 (accessed on 16 December 2021).
- Rodriguez-Morales, A.J.; Patiño-Cadavid, L.J.; Lozada-Riascos, C.O.; Villamil-Gómez, W.E. Mapping Zika in municipalities of one coastal department of Colombia (Sucre) using geographic information systems during the 2015–2016 outbreak: Implications for public health and travel advice. Int. J. Infect. Dis. 2016, 48, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Calle-Tobón, A.; Pérez-Pérez, J.; Rojo, R.; Rojas-Montoya, W.; Triana-Chavez, O.; Rúa-Uribe, G.; Gómez-Palacio, A. Surveillance of Zika virus in field-caught Aedes aegypti and Aedes albopictus suggests important role of male mosquitoes in viral populations maintenance in Medellín, Colombia. Infect. Genet. Evol. 2020, 85, 104434. [Google Scholar] [CrossRef]
- Organización Panamericana de la Salud. Actualización Epidemiológica Dengue, Chikunguña y Zika en el Contexto de COVID-19 23 de Diciembre de 2021. OPS/OMS. 2021. Available online: https://www.paho.org/es/documentos/actualizacion-epidemiologica-dengue-chikungunya-zika-contexto-covid-19-23-diciembre-2021 (accessed on 15 April 2022).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 03 de 2021. BES-Sivigila. 2021. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2021_Boletin_epidemiologico_semana_3.pdf (accessed on 27 April 2022).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 52 de 2021. BES-Sivigila. 2021. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2021_Boletin_epidemiologico_semana_52.pdf (accessed on 27 April 2022).
- Instituto Nacional de Salud. Boletín Epidemiológico Semanal (BES). Semana Epidemiológica Número 05 de 2022. BES-Sivigila. 2022. Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2022_Boletin_epidemiologico_semana_5.pdf (accessed on 27 April 2022).
- Martínez, D.; Hernández, C.; Muñoz, M.; Armesto, Y.; Cuervo, A.; Ramírez, J.D. Identification of Aedes (Diptera: Culicidae) Species and Arboviruses Circulating in Arauca, Eastern Colombia. Front. Ecol. Evol. 2020, 8, 602190. [Google Scholar] [CrossRef]
- Fernandes-Matano, L.; Monroy-Muñoz, I.E.; Pardavé-Alejandre, H.D.; Uribe-Noguez, L.A.; de los Hernández-Cueto, M.A.; Rojas-Mendoza, T.; Santacruz-Tinoco, C.E.; Grajales-Muñiz, C.; Muñoz-Medina, J.E. Impact of the introduction of chikungunya and zika viruses on the incidence of dengue in endemic zones of Mexico. PLoS Negl. Trop. Dis. 2021, 15, e0009922. [Google Scholar] [CrossRef]
- Mourya, D.; Gokhale, M.; Majumdar, T.; Yadav, P.; Kumar, V.; Mavale, M. Experimental Zika virus infection in Aedes aegypti: Susceptibility, transmission & co-infection with dengue & chikungunya viruses. Indian J. Med. Res. 2018, 147, 88–96. [Google Scholar]
- Corrêa, R.; de Oliveira Santos, I.; Braz-de-Melo, H.A.; de Sant’Ana, L.P.; das Neves Almeida, R.; Pasquarelli-do-Nascimento, G.; Prado, P.S.; Kobinger, G.P.; Maurice, C.F.; Magalhães, K.G. Gut microbiota modulation induced by Zika virus infection in immunocompetent mice. Sci. Rep. 2021, 11, 1421. [Google Scholar] [CrossRef] [PubMed]
- Bynum, N. Alpha, Beta, and Gamma Diversity. Duke University. 2021 Biology LibreTexts. 2021. Available online: https://bio.libretexts.org/@go/page/17392 (accessed on 21 November 2021).
- Dada, N.; Sheth, M.; Liebman, K.; Pinto, J.; Lenhart, A. Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep. 2018, 8, 2084. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, R.M.; Campolina, T.B.; Chaves, B.A.; Delgado, J.L.F.; Godoy, R.S.M.; Pimenta, P.F.P.; Secundino, N.F.C. The influence of culture-dependent native microbiota in Zika virus infection in Aedes aegypti. Parasites Vectors 2022, 15, 57. [Google Scholar] [CrossRef]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef]
- Ravi, A.; Halstead, F.D.; Bamford, A.; Casey, A.; Thomson, N.M.; van Schaik, W.; Snelson, C.; Goulden, R.; Foster-Nyarko, E.; Savva, G.M.; et al. Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb. Genom. 2019, 5, e000293. [Google Scholar] [CrossRef]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Rouzé, R.; Moné, A.; Delbac, F.; Belzunces, L.; Blot, N. The Honeybee Gut Microbiota is Altered after Chronic Exposure to Different Families of Insecticides and Infection by Nosema ceranae. Microb. Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Paris, L.; Peghaire, E.; Moné, A.; Diogon, M.; Debroas, D.; Delbac, F.; El Alaoui, H. Honeybee gut microbiota dysbiosis in pesticide/parasite co-exposures is mainly induced by Nosema ceranae. J. Invertebr. Pathol. 2020, 172, 107348. [Google Scholar] [CrossRef] [PubMed]
- Möhlmann, T.W.R.; Vogels, C.B.F.; Göertz, G.P.; Pijlman, G.P.; ter Braak, C.J.F.; te Beest, D.E.; Hendriks, M.; Nijhuis, E.H.; Warris, S.; Drolet, B.S.; et al. Impact of Gut Bacteria on the Infection and Transmission of Pathogenic Arboviruses by Biting Midges and Mosquitoes. Microb. Ecol. 2020, 80, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.V.; Damiani, C.; Accoti, A.; Tallarita, M.; Nunzi, E.; Cappelli, A.; Bozic, J.; Catanzani, R.; Rossi, P.; Valzano, M.; et al. Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 2018, 18, 126. [Google Scholar] [CrossRef]
- Caragata, E.P.; Otero, L.M.; Tikhe, C.V.; Barrera, R.; Dimopoulos, G. Microbial Diversity of Adult Aedes aegypti and Water Collected from Different Mosquito Aquatic Habitats in Puerto Rico. Microb. Ecol. 2021, 83, 182–201. [Google Scholar] [CrossRef]
- Duguma, D.; Hall, M.W.; Smartt, C.T.; Debboun, M.; Neufeld, J.D. Microbiota variations in Culex nigripalpus disease vector mosquito of West Nile virus and Saint Louis Encephalitis from different geographic origins. PeerJ 2019, 6, e6168. [Google Scholar] [CrossRef]
- Vijayakumar, M.M.; More, R.P.; Rangasamy, A.; Gandhi, G.R.; Muthugounder, M.; Thiruvengadam, V.; Samaddar, S.; Jalali, S.K.; Sa, T. Gut Bacterial Diversity of Insecticide-Susceptible and -Resistant Nymphs of the Brown Planthopper Nilaparvata lugens Stål (Hemiptera: Delphacidae) and Elucidation of Their Putative Functional Roles. J. Microbiol. Biotechnol. 2018, 28, 976–986. [Google Scholar] [CrossRef]
- Onyango, M.G.; Lange, R.; Bialosuknia, S.; Payne, A.; Mathias, N.; Kuo, L.; Vigneron, A.; Nag, D.; Kramer, L.D.; Ciota, A.T. Zika virus and temperature modulate Elizabethkingia anophelis in Aedes albopictus. Parasites Vectors 2021, 14, 573. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Outlaw, J.; Jaykus, L.A. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS ONE 2017, 12, e0173124. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Díaz, C.; García-Orozco, A.; Riera-Leal, A.; Padilla-Arellano, J.R.; Fafutis-Morris, M. Microbiota and Its Role on Viral Evasion: Is It with Us or Against Us? Front. Cell Infect. Microbiol. 2019, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinjé, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Almagro-Moreno, S.; Boyd, E.F. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol. Biol. 2009, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Deng, F.; Wu, S.; Wu, Y.; Liu, X.; Wu, P.; Zhai, Z. Identification of mucins and their expression in the vector mosquito Aedes albopictus. J. Vector Ecol. 2020, 45, 297–305. [Google Scholar] [CrossRef]
- Dias, R.O.; Cardoso, C.; Pimentel, A.C.; Damasceno, T.F.; Ferreira, C.; Terra, W.R. The roles of mucus-forming mucins, peritrophins and peritrophins with mucin domains in the insect midgut. Insect Mol. Biol. 2018, 27, 46–60. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Wang, L.; Guo, W.; Zhang, X.; Peng, D.; Luo, C.; Yu, Z.; Sun, M. Bacillus thuringiensis Bel Protein Enhances the Toxicity of Cry1Ac Protein to Helicoverpa armigera Larvae by Degrading Insect Intestinal Mucin. Appl. Environ. Microbiol. 2009, 75, 5237–5243. [Google Scholar] [CrossRef]
- Pelloquin, B.; Kristan, M.; Edi, C.; Meiwald, A.; Clark, E.; Jeffries, C.L.; Walker, T.; Dada, N.; Messenger, L.A. Overabundance of Asaia and Serratia Bacteria Is Associated with Deltamethrin Insecticide Susceptibility in Anopheles coluzzii from Agboville, Côte d’Ivoire. Microbiol. Spectr. 2021, 9, e00157-21. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zheng, D.; Zhong, H.; Qin, B.; Gurr, G.M.; Vasseur, L.; Lin, H.; Bai, J.; He, W.; You, M.; et al. DNA Sequencing Reveals the Midgut Microbiota of Diamondback Moth, Plutella xylostella (L.) and a Possible Relationship with Insecticide Resistance. PLoS ONE 2013, 8, e68852. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Hayatsu, M.; Hosokawa, T.; Nagayama, A.; Tago, K.; Fukatsu, T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 8618–8622. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Ma, E.; Zhu, Y.; Liu, Z.; Wei, T.; Wang, P.; Cheng, G. Interaction of Viruses with the Insect Intestine. Annu. Rev. Virol. 2021, 8, 115–131. [Google Scholar] [CrossRef]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef]
- Saraiva, R.G.; Fang, J.; Kang, S.; Angleró-Rodríguez, Y.I.; Dong, Y.; Dimopoulos, G. Aminopeptidase secreted by Chromobacterium sp. Panama inhibits dengue virus infection by degrading the E protein. PLoS Negl. Trop. Dis. 2018, 12, e0006443. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Roldán, M.D.; Pérez-Reinado, E.; Castillo, F.; Moreno-Vivián, C. Reduction of polynitroaromatic compounds: The bacterial nitroreductases. FEMS Microbiol. Rev. 2008, 32, 474–500. [Google Scholar] [CrossRef] [PubMed]
- Boddu, R.S.; Perumal, O.; Divakar, K. Microbial nitroreductases: A versatile tool for biomedical and environmental applications. Biotechnol. Appl. Biochem. 2020, 68, 1518–1530. [Google Scholar] [CrossRef]
- Petersen, L.M.; Tisa, L.S. Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 2013, 59, 627–640. [Google Scholar] [CrossRef]
- Lerouge, I.; Vanderleyden, J. O-antigen structural variation: Mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol. Rev. 2002, 26, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, J.N.; Kinsinger, N.; Perring, T.M.; Backus, E.A.; Shugart, H.J.; Walker, S.; Roper, M.C. O Antigen Modulates Insect Vector Acquisition of the Bacterial Plant Pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 2015, 81, 8145–8154. [Google Scholar] [CrossRef] [PubMed]
- Van Houdt, R.; Givskov, M.; Michiels, C.W. Quorum sensing in Serratia. FEMS Microbiol. Rev. 2007, 31, 407–424. [Google Scholar] [CrossRef]
- Chen, J.; Xie, J. Role and regulation of bacterial LuxR-like regulators. J. Cell Biochem. 2011, 112, 2694–2702. [Google Scholar] [CrossRef]
- Reeves, P.R.; Cunneen, M.M. Biosynthesis of O-antigen chains and assembly. In Microbial Glycobiology; Elsevier: Amsterdam, The Netherlands, 2010; pp. 319–335. [Google Scholar]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; Van Breusegem, F.; Eberl, L.; et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef]
- Suryawanshi, R.K.; Koujah, L.; Patil, C.D.; Ames, J.M.; Agelidis, A.; Yadavalli, T.; Patil, S.V.; Shukla, D. Bacterial Pigment Prodigiosin Demonstrates a Unique Antiherpesvirus Activity That Is Mediated through Inhibition of Prosurvival Signal Transducers. J. Virol. 2020, 94, e00251-20. [Google Scholar] [CrossRef]
- Da Mota, F.F.; Castro, D.P.; Vieira, C.S.; Gumiel, M.; de Albuquerque, J.P.; Carels, N.; Zambuja, P. In vitro Trypanocidal Activity, Genomic Analysis of Isolates, and in vivo Transcription of Type VI Secretion System of Serratia marcescens Belonging to the Microbiota of Rhodnius prolixus Digestive Tract. Front. Microbiol. 2018, 9, 3205. [Google Scholar] [CrossRef]
- Parvin, W.; Govender, N.; Othman, R.; Jaafar, H.; Rahman, M.; Wong, M.Y. Phenazine from Pseudomonas aeruginosa UPMP3 induced the host resistance in oil palm (Elaeis guineensis Jacq.)-Ganoderma boninense pathosystem. Sci. Rep. 2020, 10, 15621. [Google Scholar] [CrossRef] [PubMed]
- Dar, D.; Thomashow, L.S.; Weller, D.M.; Newman, D.K. Global landscape of phenazine biosynthesis and biodegradation reveals species-specific colonization patterns in agricultural soils and crop microbiomes. eLife 2020, 9, e59726. [Google Scholar] [CrossRef] [PubMed]
- Crotti, E.; Damiani, C.; Pajoro, M.; Gonella, E.; Rizzi, A.; Ricci, I.; Negri, I.; Scuppa, P.; Rossi, P.; Ballarini, P.; et al. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 2009, 11, 3252–3264. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Ricci, I.; Crotti, E.; Rossi, P.; Rizzi, A.; Scuppa, P.; Capone, A.; Ulissi, U.; Epis, S.; Genchi, M.; et al. Mosquito-Bacteria Symbiosis: The Case of Anopheles gambiae and Asaia. Microb. Ecol. 2010, 60, 644–654. [Google Scholar] [CrossRef]
- Cappelli, A.; Damiani, C.; Mancini, M.V.; Valzano, M.; Rossi, P.; Serrao, A.; Ricci, I.; Favia, G. Asaia Activates Immune Genes in Mosquito Eliciting an Anti-Plasmodium Response: Implications in Malaria Control. Front. Genet. 2019, 10, 836. [Google Scholar] [CrossRef]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef]
- Rossi, P.; Ricci, I.; Cappelli, A.; Damiani, C.; Ulissi, U.; Mancini, M.V.; Valzano, M.; Capone, A.; Epis, S.; Crotti, E.; et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasites Vectors 2015, 8, 278. [Google Scholar] [CrossRef]
- Brinzer, R.A.; Henderson, L.; Marchiondo, A.A.; Woods, D.J.; Davies, S.A.; Dow, J.A.T. Metabolomic profiling of permethrin-treated Drosophila melanogaster identifies a role for tryptophan catabolism in insecticide survival. Insect Biochem. Mol. Biol. 2015, 67, 74–86. [Google Scholar] [CrossRef]
- Feng, Y.; Peng, Y.; Wen, H.; Song, X.; An, Y.; Tang, H.; Wang, J. Microbial tryptophan catabolism affects the vector competence of Anopheles. Microbiology, 2021; preprint. Available online: http://biorxiv.org/lookup/doi/10.1101/2021.02.15.431262 (accessed on 1 March 2022).
- Dintner, S.; Staroń, A.; Berchtold, E.; Petri, T.; Mascher, T.; Gebhard, S. Coevolution of ABC Transporters and Two-Component Regulatory Systems as Resistance Modules against Antimicrobial Peptides in Firmicutes Bacteria. J. Bacteriol. 2011, 193, 3851–3862. [Google Scholar] [CrossRef]
- Meehl, M.; Herbert, S.; Götz, F.; Cheung, A. Interaction of the GraRS Two-Component System with the VraFG ABC Transporter to Support Vancomycin-Intermediate Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef]
- Ahmad, A.; Majaz, S.; Nouroz, F. Two-component systems regulate ABC transporters in antimicrobial peptide production, immunity and resistance. Microbiology 2020, 166, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Epis, S.; Porretta, D.; Mastrantonio, V.; Comandatore, F.; Sassera, D.; Rossi, P.; Cafarchia, C.; Otranto, D.; Favia, G.; Genchi, C.; et al. ABC transporters are involved in defense against permethrin insecticide in the malaria vector Anopheles stephensi. Parasites Vectors 2014, 7, 349. [Google Scholar] [CrossRef] [PubMed]
- Sheets, J.J.; Hey, T.D.; Fencil, K.J.; Burton, S.L.; Ni, W.; Lang, A.E.; Benz, R.; Aktories, K. Insecticidal Toxin Complex Proteins from Xenorhabdus nematophilus. J. Biol. Chem. 2011, 286, 22742–22749. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, W.J.; Pilz-Júnior, H.L.; Heermann, R.; da Silva, O.S. The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: A review. Parasites Vectors 2020, 13, 376. [Google Scholar] [CrossRef]
- Yooyangket, T.; Muangpat, P.; Polseela, R.; Tandhavanant, S.; Thanwisai, A.; Vitta, A. Identification of entomopathogenic nematodes and symbiotic bacteria from Nam Nao National Park in Thailand and larvicidal activity of symbiotic bacteria against Aedes aegypti and Aedes albopictus. PLoS ONE 2018, 13, e0195681. [Google Scholar] [CrossRef]
- Park, Y.; Kim, Y. Eicosanoids rescue Spodoptera exigua infected with Xenorhabdus nematophilus, the symbiotic bacteria to the entomopathogenic nematode Steinernema carpocapsae. J. Insect Physiol. 2000, 46, 1469–1476. Available online: https://pubmed.ncbi.nlm.nih.gov/10891575/ (accessed on 18 July 2022). [CrossRef]
- Eom, S.; Park, Y.; Kim, Y. Sequential immunosuppressive activities of bacterial secondary metabolites from the entomopahogenic bacterium Xenorhabdus nematophila. J. Microbiol. 2014, 52, 161–168. [Google Scholar] [CrossRef]
- Colclough, A.L.; Scadden, J.; Blair, J.M.A. TetR-family transcription factors in Gram-negative bacteria: Conservation, variation and implications for efflux-mediated antimicrobial resistance. BMC Genom. 2019, 20, 731. [Google Scholar] [CrossRef]
- Porretta, D.; Gargani, M.; Bellini, R.; Medici, A.; Punelli, F.; Urbanelli, S. Defence mechanisms against insecticides temephos and diflubenzuron in the mosquito Aedes caspius: The P-glycoprotein efflux pumps. Med. Vet. Entomol. 2008, 22, 48–54. [Google Scholar] [CrossRef]
- Tay, J.W.; Choe, D.H.; Mulchandani, A.; Rust, M.K. Hydrogels: From Controlled Release to a New Bait Delivery for Insect Pest Management. J. Econ. Entomol. 2020, 113, 2061–2068. [Google Scholar] [CrossRef]
- Roy, A.; Singh, S.K.; Bajpai, J.; Bajpai, A.K. Controlled pesticide release from biodegradable polymers. Cent. Eur. J. Chem. 2014, 12, 453–469. [Google Scholar] [CrossRef]
- Djiappi-Tchamen, B.; Nana-Ndjangwo, M.S.; Nchoutpouen, E.; Makoudjou, I.; Ngangue-Siewe, I.N.; Talipouo, A.; Mayi, M.P.A.; Awono-Ambene, P.; Wondji, C.; Tchuinkam, T.; et al. Aedes Mosquito Surveillance Using Ovitraps, Sweep Nets, and Biogent Traps in the City of Yaoundé, Cameroon. Insects 2022, 13, 793. [Google Scholar] [CrossRef] [PubMed]
- Zouache, K.; Raharimalala, F.N.; Raquin, V.; Tran-Van, V.; Raveloson, L.H.R.; Ravelonandro, P.; Mavingui, P. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 2011, 75, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Gómez-Martínez, C.; Chin, Y.; Saltonstall, K.; McMillan, W.O.; Rovira, J.R.; Loaiza, J.R. Dynamics and diversity of bacteria associated with the disease vectors Aedes aegypti and Aedes albopictus. Sci. Rep. 2019, 9, 12160. [Google Scholar] [CrossRef]
- Rani, A.; Sharma, A.; Rajagopal, R.; Adak, T.; Bhatnagar, R.K. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol. 2009, 9, 96. [Google Scholar] [CrossRef]
- Yadav, K.K.; Bora, A.; Datta, S.; Chandel, K.; Gogoi, H.K.; Prasad, G.B.K.S.; Veer, V. Molecular characterization of midgut microbiota of Aedes albopictus and Aedes aegypti from Arunachal Pradesh, India. Parasit Vectors 2015, 8, 641. [Google Scholar] [CrossRef]
- Scolari, F.; Casiraghi, M.; Bonizzoni, M. Aedes spp. and Their Microbiota: A Review. Front. Microbiol. 2019, 10, 2036. [Google Scholar] [CrossRef]
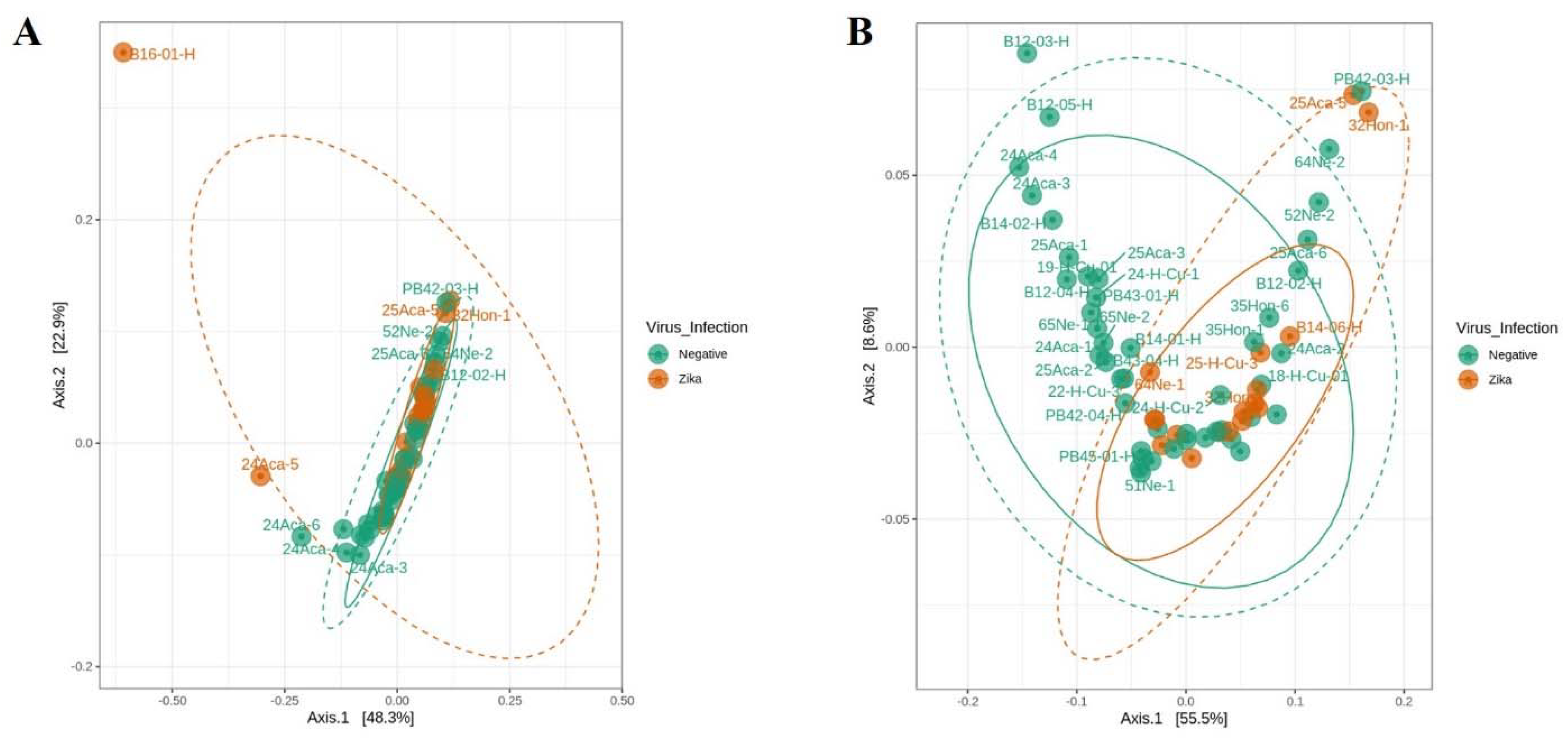


| Degrees of Freedom | Sum of Squares | R2 | F | Pr (>F) | |
|---|---|---|---|---|---|
| ZIKV infection | 1 | 0.0483 | 0.0666 | 4.2839 | 0.0096 * |
| Residual | 60 | 0.6768 | 0.9334 | ||
| Total | 61 | 0.7252 | 1.000 |
| Degrees of Freedom | Sum of Squares | R2 | F | Pr (>F) | |
|---|---|---|---|---|---|
| lambda-cyhalothrin resistance | 2 | 0.0175 | 0.0242 | 0.7313 | 0.6113 |
| Residual | 59 | 0.7076 | 0.9758 | ||
| Total | 61 | 0.7252 | 1.000 |
| Degrees of Freedom | Sum of Squares | R2 | F | Pr (>F) | |
|---|---|---|---|---|---|
| Virus infection | 1 | 0.0528 | 0.0728 | 4.7092 | 0.0063 * |
| lambda-cyhalothrin resistance | 1 | 0.0156 | 0.0215 | 1.3882 | 0.1989 |
| Residual | 59 | 0.6613 | 0.9119 | ||
| Total | 61 | 0.7252 | 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arévalo-Cortés, A.; Damania, A.; Granada, Y.; Zuluaga, S.; Mejia, R.; Triana-Chavez, O. Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations. Viruses 2022, 14, 2197. https://doi.org/10.3390/v14102197
Arévalo-Cortés A, Damania A, Granada Y, Zuluaga S, Mejia R, Triana-Chavez O. Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations. Viruses. 2022; 14(10):2197. https://doi.org/10.3390/v14102197
Chicago/Turabian StyleArévalo-Cortés, Andrea, Ashish Damania, Yurany Granada, Sara Zuluaga, Rojelio Mejia, and Omar Triana-Chavez. 2022. "Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations" Viruses 14, no. 10: 2197. https://doi.org/10.3390/v14102197
APA StyleArévalo-Cortés, A., Damania, A., Granada, Y., Zuluaga, S., Mejia, R., & Triana-Chavez, O. (2022). Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations. Viruses, 14(10), 2197. https://doi.org/10.3390/v14102197







